Fig. 14.1
Block diagram of the EEG-ECG based online seizure detection architecture
The data captured from the 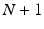 sensors (where N are the EEG electrodes plus one ECG channel) have been synchronized and transmitted as streams of multidimensional signals. Thus, the input to the illustrated in Fig. 14.1 architecture consists of time-synchronous streams of EEG and ECG signal samples. As shown in Fig. 14.1, in a first step the EEG,
sensors (where N are the EEG electrodes plus one ECG channel) have been synchronized and transmitted as streams of multidimensional signals. Thus, the input to the illustrated in Fig. 14.1 architecture consists of time-synchronous streams of EEG and ECG signal samples. As shown in Fig. 14.1, in a first step the EEG, 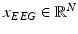 , and ECG,
, and ECG, 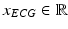 , signals are pre-processed. Pre-processing consists of frame blocking of the incoming streams to epochs of constant length w with constant time-shift s. Each epoch is a
, signals are pre-processed. Pre-processing consists of frame blocking of the incoming streams to epochs of constant length w with constant time-shift s. Each epoch is a 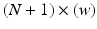 matrix, where N is the number of EEG electrodes and
matrix, where N is the number of EEG electrodes and 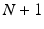 is the N-dimensional EEG signal appended by the ECG signal.
is the N-dimensional EEG signal appended by the ECG signal.
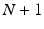 sensors (where N are the EEG electrodes plus one ECG channel) have been synchronized and transmitted as streams of multidimensional signals. Thus, the input to the illustrated in Fig. 14.1 architecture consists of time-synchronous streams of EEG and ECG signal samples. As shown in Fig. 14.1, in a first step the EEG,
sensors (where N are the EEG electrodes plus one ECG channel) have been synchronized and transmitted as streams of multidimensional signals. Thus, the input to the illustrated in Fig. 14.1 architecture consists of time-synchronous streams of EEG and ECG signal samples. As shown in Fig. 14.1, in a first step the EEG, 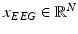 , and ECG,
, and ECG, 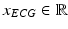 , signals are pre-processed. Pre-processing consists of frame blocking of the incoming streams to epochs of constant length w with constant time-shift s. Each epoch is a
, signals are pre-processed. Pre-processing consists of frame blocking of the incoming streams to epochs of constant length w with constant time-shift s. Each epoch is a 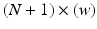 matrix, where N is the number of EEG electrodes and
matrix, where N is the number of EEG electrodes and 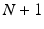 is the N-dimensional EEG signal appended by the ECG signal.
is the N-dimensional EEG signal appended by the ECG signal.After pre-processing, the extracted epochs are in parallel processed by time-domain and frequency-domain feature extraction algorithms separately for the N-dimensional EEG and the 1-dimensional ECG signals. In particular, each of the N-dimensions of the EEG signal are processed by time-domain and frequency-domain feature extraction algorithms for EEG, while the ECG signal is processed by time-domain feature extraction algorithms (based on heart rate estimation) dedicated for electrocardiogram, as shown in the block diagram of Fig. 14.1. The extracted time-domain and frequency-domain features for the EEG, 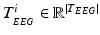 and
and 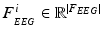 , with
, with 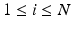 , and the ECG signal,
, and the ECG signal, 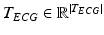 , are afterwards concatenated to a single feature vector
, are afterwards concatenated to a single feature vector 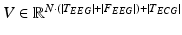 representing each epoch, as shown in Fig. 14.1. The extracted sequences of feature vectors, V, are short-time parametric representations of the EEG and ECG signals representing the time and spectral characteristics of the multimodal signals. This sequence of feature vectors is afterwards used as input to a classification model in order to assign a class label (seizure class or non-seizure class) to each of the vectors, i.e. to the corresponding time-intervals of the vectors.
representing each epoch, as shown in Fig. 14.1. The extracted sequences of feature vectors, V, are short-time parametric representations of the EEG and ECG signals representing the time and spectral characteristics of the multimodal signals. This sequence of feature vectors is afterwards used as input to a classification model in order to assign a class label (seizure class or non-seizure class) to each of the vectors, i.e. to the corresponding time-intervals of the vectors.
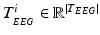 and
and 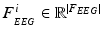 , with
, with 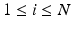 , and the ECG signal,
, and the ECG signal, 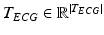 , are afterwards concatenated to a single feature vector
, are afterwards concatenated to a single feature vector 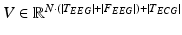 representing each epoch, as shown in Fig. 14.1. The extracted sequences of feature vectors, V, are short-time parametric representations of the EEG and ECG signals representing the time and spectral characteristics of the multimodal signals. This sequence of feature vectors is afterwards used as input to a classification model in order to assign a class label (seizure class or non-seizure class) to each of the vectors, i.e. to the corresponding time-intervals of the vectors.
representing each epoch, as shown in Fig. 14.1. The extracted sequences of feature vectors, V, are short-time parametric representations of the EEG and ECG signals representing the time and spectral characteristics of the multimodal signals. This sequence of feature vectors is afterwards used as input to a classification model in order to assign a class label (seizure class or non-seizure class) to each of the vectors, i.e. to the corresponding time-intervals of the vectors.During the training phase of the seizure detector, a dataset of feature vectors with known class labels (labeled manually by medical experts) is used to train a binary model M (two classes: seizure vs. non-seizure) using a classification algorithm f. At the test phase the existing seizure model, M, is used in order to decide for each epoch’s feature vector, V, the corresponding class using the same classification algorithm, f, as in the training phase. Thus, for each epoch i a binary label d i , i.e. seizure or not, is decided as:
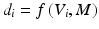
and the sequence of incoming EEG-ECG data is decomposed to time-intervals of seizure or clear (non-seizure) recordings. Post-processing of the automatically detected labels can be performed for improving the performance of the architecture.
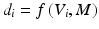
(1)
During pre-processing the time-synchronized EEG and ECG recordings were frame blocked to epochs of 1-s length, without time-overlap between successive epochs. For each epoch, time-domain and frequency domain features were extracted separately for the each of the 21 EEG channels and the ECG channel.
In particular, each of the EEG channels was parameterized using the following features: time-domain features: minimum value, maximum value, mean, variance, standard deviation, percentiles (25 %, 50 %-median and 75 %), interquartile range, mean absolute deviation, range, skewness, kurtosis, energy, Shannon’s entropy, logarithmic energy entropy, number of positive and negative peaks, zero-crossing rate, and frequency-domain features: Sixth order autoregressive-filter (AR) coefficients, power spectral density, frequency with maximum and minimum amplitude, spectral entropy, delta-theta-alpha-beta-gamma band energy, discrete wavelet transform coefficients with mother wavelet function Daubechies 16 and decomposition level equal to eight, thus resulting to a feature vector of dimensionality equal to 55 for each of the 21 EEG channels, i.e. 1155 in total.
The ECG channel was parameterized using the following features: the heart rate absolute value and variability statistics of the heart rate, i.e. minimum value, maximum value, mean, variance, standard deviation, percentiles (25 %, 50 %-median and 75 %), interquartile range, mean absolute deviation, range, thus resulting to a feature vector of dimensionality equal to 12. The heart rate estimation was based on Shannon energy envelope estimation for R-peak detection algorithm, implemented as in [2]. The dimensionality of the overall feature vector V is 1155 + 12 = 1167.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree








