From Genes to Brain: Developmental Neurobiology
James F. Leckman
Flora M. Vaccarino
Paul J. Lombroso
Human beings are complex living organisms that can be characterized by their appearance and behavior at each point in their life cycle. Many of these characteristics are uniquely human, such as the array of languages that facilitate interpersonal communication and permit a meaningful interplay of ideas and emotions. Other characteristics, such as affection and aggression, are less distinctive and place our species as one among many that populate the earth. Scientific advances over the past 150 years clearly indicate that hereditary factors that are transmitted from generation to generation account for much of the observed variation among and within species. Although the complexities of human existence cannot be reduced simply to the effects of genes, it is inescapable that genetic factors provide the biological basis for many of our potentialities and vulnerabilities as human beings (1).
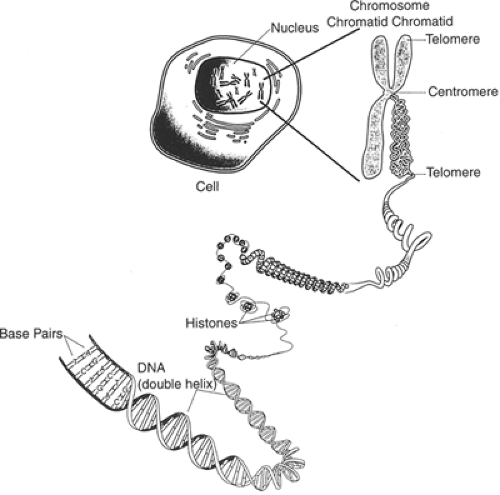
Our genetic endowment, as a species, is a unique collection of discrete units of heredity (genes) that for the most part are linearly arranged on 46 chromosomes (22 pairs of homologous chromosomes and two sex chromosomes) (Figure 2.3.1.1). It is apparent from the completion of the human genome sequencing project that there are approximately 35,000 genes on our chromosomes. This collection of genes makes us both alike and different from other organisms. Although the precise genetic determinants of our interspecies similarities and differences are largely obscure, it is probable that many of the responsible genetic factors will be identified. For example, investigators are in the midst of discovering the cascade of genes that have contributed to the remarkable neuroanatomical and functional evolution of the cerebral neocortices across different mammalian species over the past 50 million years (2).
Genetic factors also contribute to variations within species. A large number of physical and psychological traits, including gender, height, and intelligence, have been shown to be under at least partial genetic control. One need only examine the striking physical and psychological similarities between monozygotic (genetically identical) twins reared apart to recognize the powerful influence of genes in determining who we are. (3,4) Some of these intraspecies differences, such as gender, are due to actual differences in the number and type of genes present in the individual. For example, genes on the Y chromosome are present in males and are transmitted from father to son. Other intraspecies differences are due to multiple forms (polymorphic alleles) of specific genes that are distributed within the population. The polymorphisms are due to differences in the basic elements of genes, namely the linear sequence of nucleotides that form our chromosomes. For the most part, these polymorphisms are silent in the sense that they do not change the proteins that are encoded. However, some of the changes lead to differences in the structure, activity or level of expression of proteins, which may then contribute to traits such as blood type, height, or eye color. In others, the changes within the genes are subtler, and make the individual more susceptible to additional factors, be they genetic or environmental, that may lead to phenotypic expression of clinical significance.
Some allelic variations are so significant that we use the term mutation to signify that the changes will usually lead to disease states such as Rett syndrome, Huntington’s disease, Marfan syndrome, or sickle cell anemia, disorders in which other ameliorating factors will have little effect. Finally, some intraspecies differences may depend on which parent passed on a particular piece of genetic material, a process termed genetic imprinting. (5) Briefly, imprinting affects the structure of the deoxyribonucleic acid (DNA) molecule but not the nucleotide sequence itself. Mechanisms that modify the DNA structure without changing the nucleotide sequence are termed epigenetic phenomena. For example, proteins that regulate gene expression are not able to gain access to imprinted genes, and genes within an imprinted chromosomal region are effectively silenced. The most dramatic example of imprinting concerns two distinctively different developmental disorders, Prader-Willi and Angelman’s syndrome, that are due to alterations of DNA in the same general chromosomal region on chromosome 15 (6).
Apart from interspecies and intraspecies variations, it is also important to recognize that differences over the life span of an individual member of a species can also be due to genetic factors. Simply put, this means that not all genes are active at the same time. For example, the hemoglobin genes active during fetal life are different from those that are active in adulthood. Differences in the pattern of expression of genes over the life span are largely responsible for the physical and cognitive functioning of individual organisms, from Drosophila to humans. These differences are also responsible for the allocation of specific characteristics and functions to specific cells of the body, e.g., for making muscle cells different from brain cells, and are orchestrated by master developmental genes in a precise manner. As the temporal patterns of gene expression during human development become known, we may come to be able to better predict each individual’s genetic potential.
Genes
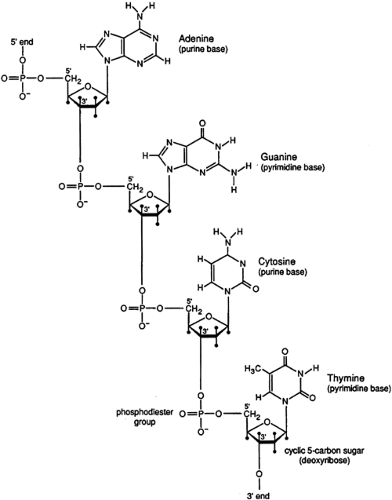
Mendel first postulated the existence of discrete hereditary factors or genes in 1865 (9) but their importance was not appreciated until the early 1900s. As indicated above, genes are arranged linearly on chromosomes that are found in the nuclei of most cells (Figure 2.3.1.2). They are composed of DNA. DNA consists of a string of nucleotides (complex molecules that contain a sugar moiety, a phosphate group, and either a purine or a pyrimidine base) that are linked together. These linkages involve connecting the carbon atom of one sugar moiety to the carbon atom of the next nucleotide through a phosphate group. Four separate nucleotides are found in DNA. Two contain purine bases (adenine and guanine) and two contain pyrimidine bases (thymine and cytosine).
Genetic information is conveyed by the nucleotide sequence of a particular DNA molecule. Most DNA molecules exist in a double helical structure composed of two polynucleotide chains held together by a series of hydrogen bonds between complementary base pairs (adenine is bonded to thymine and guanine to cytosine) (Figure 2.3.1.3). This structure confers stability of the molecule and provides the basis for replication. Since each strand of DNA in the double helix is exactly complementary to the other, knowing the sequence of one strand provides precise knowledge of the sequence of the other.
The sequence of nucleotides on a DNA molecule determines the order of the 20 different amino acids in proteins. As a consequence, the information contained in DNA provides the instructions for cells to grow and divide, set in motion developmental sequences that lead to orderly differentiation
of cell types, and provide for the maintenance of a diversified population of cells that are necessary for the successful functioning of complex organisms.
of cell types, and provide for the maintenance of a diversified population of cells that are necessary for the successful functioning of complex organisms.
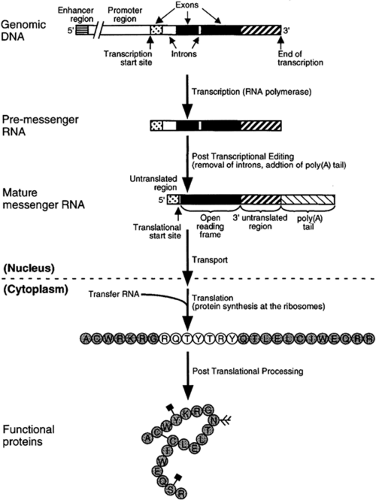
The nucleotide sequence, however, does not provide a direct template for protein synthesis. Instead, there are a series of intervening steps that require the DNA to be transcribed into messenger ribonucleic acid (mRNA). Messenger RNA molecules are very similar in composition to DNA and can hybridize with complementary DNA sequences. Mature mRNA molecules are rapidly transported out of the nucleus, where they serve as the template for protein synthesis.
The translation of a message into a specific amino acid sequence occurs at ribosomes located in either the cytoplasm or attached to the endoplasmic reticulum. The amino acid sequence is determined by the sequence of bases, with sets of three bases constituting a codon that stands for an amino acid. At the ribosome, codons of an mRNA molecule bind to complementary anticodons of transfer RNAs (tRNAs), which then transfer specific amino acids to the growing protein chain. The basic elements of this “central dogma” of protein production were first proposed by Crick (10).
Genes are normally extremely stable and are precisely copied during the chromosomal duplications that precede cell divisions (mitosis). Obviously, any mistakes that occur have the potential of disturbing the normal sequence of amino acids in a protein. There are a number of proteins within the nucleus whose function it is to recognize and repair errors within the DNA sequence. Very rarely, however, mistakes go uncorrected and will result in changes in the original nucleotide sequence. The majority of such changes have no effects as they occur in regions on the DNA molecule that do not encode for protein. However, mutations that occur within the sequence that encodes for protein or within regulatory portions of genes may have functional consequences. First, they may slightly change the enzymatic function of the protein, leading to the allelic variations discussed above. Second, when a critical amino acid is mutated, there may be dramatic changes in the function of the encoded protein and, ultimately, to the organism. Finally, this capacity for change can, in rare instances, lead to positive consequences that serve as the basis for evolution.
Regulation of Gene Function
According to some estimates, only about 1 percent of the genome is being expressed at a given time in higher eukaryotic cells (8). Different sets of genes are active during development compared to the mature organism, and different genes are required in the various tissues. Some genes are expressed continuously, while others are required during specific periods, and others are only expressed in response to hormonal or environmentally triggered changes. Regulation of gene function can occur at any of the many steps required for gene expression (Figure 2.3.1.4).
Transcriptional Factors
The basic flow of biological information is from DNA to RNA (transcription) and from RNA to protein (translation). Transcription of mRNA is initiated by the binding of a protein complex containing RNA polymerase II to the regulatory region of a gene, termed the promoter. These regions are usually found immediately upstream of the transcriptional start site. Sequences of highly conserved nucleotides within the
promoter serve as binding sites for the regulatory proteins required to initiate transcription. After binding, the two DNA strands unravel and the sense strand is accessible to the polymerase enzyme. In addition, other highly conserved DNA sequences have been identified that either enhance or repress the transcription of target genes. These sequences are usually found near the regulatory promoter region or in further upstream regions of DNA. Enhancer or repressor sequences bind regulatory proteins called transcription factors to form complexes that allow the RNA polymerase II complex to bind more or less efficiently to the underlying gene. In this way, specific sets of genes are expressed while others are repressed, depending on the precise mixture of enhancers, repressors, and transcription factors present in the cell. It should be pointed out that mutations that occur within the promoter regions of genes can have a dramatic effect on the expression of the encoded protein by disrupting the binding sites of the basal transcriptional machinery, repressors, or enhancer proteins.
promoter serve as binding sites for the regulatory proteins required to initiate transcription. After binding, the two DNA strands unravel and the sense strand is accessible to the polymerase enzyme. In addition, other highly conserved DNA sequences have been identified that either enhance or repress the transcription of target genes. These sequences are usually found near the regulatory promoter region or in further upstream regions of DNA. Enhancer or repressor sequences bind regulatory proteins called transcription factors to form complexes that allow the RNA polymerase II complex to bind more or less efficiently to the underlying gene. In this way, specific sets of genes are expressed while others are repressed, depending on the precise mixture of enhancers, repressors, and transcription factors present in the cell. It should be pointed out that mutations that occur within the promoter regions of genes can have a dramatic effect on the expression of the encoded protein by disrupting the binding sites of the basal transcriptional machinery, repressors, or enhancer proteins.
Some of these highly conserved repetitive DNA sequences in the promoter regions of genes serve as phylogenetic “footprints,” which are reliable guides to regulatory regions of genes (11). As such, their presence is being used as one element of algorithms to identify putative novel genes within the vast sequence data bases generated by the human genome project and related commercial efforts (12).
Several classes of DNA-binding proteins exist and regulate the transcription of most genes. The best characterized of these transcription factors contain conserved amino acid regions that bind to specific DNA sequences. For example, homeodomain transcription factors have a highly conserved 60-amino acid region with a helix-turn-helix structure. This structure contains two helices that bind a specific DNA sequence in the major groove of the double helix of DNA (13). Other transcriptional factors have different tertiary structures known as zinc fingers and leucine zippers that also bind to DNA regulatory sequences within the promoter and regulate the initiation of transcription (14,15).
Another major issue concerns the mechanisms by which genes remain quiescent in some cell lineages or are repressed after a period of activity (16). This is a critical issue during the complex cascade in time and space of the genes necessary for the formation of the brain. During specific points in development, different genes are sequestered within heritable forms of chromatin complex that preclude transcription. Developmental repression of adult gene expression is remarkably efficient. Ratios of 1:10,000,000 or more have been estimated for the level of globin or growth hormone transcripts in cells not expressing these genes compared with those that are (17,18).
One way to achieve gene repression is for transcription factors to modify the structure of the DNA molecule itself.
One such process is through the acetylation of histones. In normal transcription, histone acetylation causes an unwinding of the DNA that permits the transcriptional machinery to bind to the promoter region. Consequently, factors that activate histone acetylases promote gene transcription.
One such process is through the acetylation of histones. In normal transcription, histone acetylation causes an unwinding of the DNA that permits the transcriptional machinery to bind to the promoter region. Consequently, factors that activate histone acetylases promote gene transcription.
Another important mechanism of gene regulation is achieved through CpG islands that are present within the regulatory regions of most genes. “CpG islands” refer to the presence of nucleotide sequences rich in C + G nucleotides. The protein methyl-CpG binding protein (Mecp2) is thought to bind to methylated CpG dinucleotides in the mammalian genome and to recruit the repressor Sin3A and histone deacetylase (Hdac) (19,20,21). In addition, Mecp2 associates with both histone H2 Lys9 methyltransferase and with the DNA methyltransferase Dnmt1 (22,23), which are potent inhibitors of transcription. Intriguingly, mutations in Mecp2 have been identified in most individuals with Rett syndrome (24). Repression of transcription via Mecp2 and associated proteins has emerged as being an important factor in two neurodevelopmental disorders, Rett and fragile X syndrome (25) (see Chapter 2.3.3, Lombroso, for a more detailed examination of these two syndromes), as well as the molecular events leading to various cancers.
DNA in somatic tissue is characterized by a bimodal pattern of methylation, which is established through a series of developmental events (26). Very early in development, most DNA is unmethylated, but after implantation, a wave of de novo methylation modifies most of the genome, excluding the majority of CpG islands, which are mainly associated with housekeeping genes. These genomic methylation patterns are broadly maintained during the life of the organism by maintenance methylation and generally correlate with gene expression.
“Gene dosage” is another crucial issue in the regulation of genes. Under normal circumstances in adults there are classes of genes where only one of the two inherited copies is active. Two prominent examples include genes that are not expressed due to the early coordinated inactivation of one copy of the two X chromosomes in women, and those genes that are not expressed due to genomic imprinting. Imprinting involves the coordinated silencing of contiguous genes coming either from the father or mother. Although the mechanisms that underlie such events are poorly understood, significant progress is being made to understand these processes at the molecular level (27,28,29).
It is evident from this description that gene transcription depends on a complex combination of regulatory mechanisms. A different pattern of gene expression will emerge depending on which nucleotide sequences are present within a promoter region as well as which transcription factors, enhancers, or repressor proteins are present within the cell. Moreover, this interplay of regulatory factors will determine whether and how much of a specific protein is transcribed. We will return to these concepts below when we describe exactly how certain growth factors as well as environmental factors interplay with transcription factors to either initiate or repress expression of genes within the brain.
A species’ genetic program unfolds in a largely predictable fashion despite its formidable complexity from the earliest gene expression in the zygote through the entire morphogenesis of the organism. This uniformity may depend, in part, on the presence of redundant pathways and in part from the fact that development proceeds largely in the direction of increasing complexity, but lesser potential.
Posttranscriptional Events
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree