Fig. 7.1
(a) Coronal MR image through occipital lobe, including calcarine sulcus. Normal human subject. MT-weighted Turbo Spin Echo (TSE) sequence, 7 T MR scanner (MAGNETOM 7 T, Siemens). Voxel size is (0.50 mm)3. Acquisition time was 14 min 33 s for 20 slices. (b) Expanded view of box in Fig. 7.1a. (c) Comparable section of lower bank of calcarine sulcus in cadaver human brain. Merker stained (cell body). (d) Adjacent section using Gallyas myelin stain, showing Stria of Gennari in cortical layer IV, corresponding closely in appearance and location to the in-vivo MR images in (a) and (b)
It is thus important to map MR-accessible parameters that correlate well with the cortical myelin content (Hopf 1954, 1955, 1956) measured in cadaver brain sections. Among suggested candidates are proton density (Shah et al. 2011), myelin water fraction (MWF, Mackay et al. 2006), magnetization transfer rate (MTR, Wolff and Balaban 1989; Koenig 1991), transverse relaxation rate (R2*, Duyn 2011), phase maps formed using gradient echo imaging (Duyn et al. 2007), and longitudinal relaxation rate (R1) (Fischl et al. 2004). The challenge is to obtain a high enough spatial resolution to distinguish intra-cortical details that can assist in realistic parcellation, within a scanning time short enough for in-vivo studies of humans.
At this point it must be mentioned that currently there is no technique in existence that can quantitatively assay the density of myelin at high spatial resolution in cadaver brain tissue. The optical density of myelin stained sections of cadaver brain clearly reflects the amount of myelin, but this does not correspond to actual myelin density, because the microscope images are essentially shadow-grams, like flat X-rays, giving no information about the distribution of myelin in the third dimension perpendicular to the plane of the section. Chemical assays, such as those pioneered in the nineteenth century by Thudichum (1884), require volumes of tissue, at least 30 mm3, that preclude intracortical spatial resolution (Dasgupta and Hogan 2001).
Because the water content of grey and white brain tissue is about 80 % and 70 % respectively, it is possible to obtain useful contrast simply using maps of proton density (Shah et al. 2011), which demonstrate this very basic physical distinction. These can be obtained by means of very short echo time (TE) imaging, or by extrapolating sets of images with a range of echo times to zero TE. However, high signal to noise ratio (SNR) is required because of the relatively small grey-white difference in water content, and this method has not been widely used for distinguishing intra-cortical structures.
Myelin water fraction (MWF) is defined as the fraction of the NMR signal from water protons that can be unambiguously assigned to the thin layers of water that separate the layers of myelin membrane that wrap myelinated axons. There is good evidence (Kalantari et al. 2011) that this water exchanges very slowly with the other major pools of water in brain tissue, those within cell bodies and between cells. MWF can be estimated by determining the distribution of T2 relaxation times (Whittall et al. 1997). In brain tissue, at 3 T magnetic field strength, T2 distributions show three peaks, empirically assigned to compartmentalized spin populations: a short T2 peak around 20 ms (between 10 and 50 ms) representing the water trapped between the layers of myelin, a second peak around 70–90 ms assigned to intra- and extracellular water, and a third peak with T2 > 2 s usually assigned to cerebrospinal fluid signal. The myelin water fraction can be computed as the fraction of signal in the T2 distribution below about 40 ms, typically representing about 15 % of the total water content. T2 relaxation times can be obtained using the well-known Carr-Purcell-Meiboom-Gill technique (Kolind et al. 2009).
There are two practical limitations regarding the use of MWF for investigation of cortical myeloarchitecture. Firstly, as just mentioned, because there is no simple quantitative assay for myelin content in living human brain, it has not yet been demonstrated that MWF provides a linear measure of myelin density. The second limitation is the comparatively low signal-to-noise ratio (SNR) of accurate MWF methods, which means that either very large voxels or very long averaging times must be used. Although a faster sequence (mcDESPOT) based on steady-state free precession and fast gradient echo acquisition has been developed (Deoni et al. 2008), this method for quantifying MWF is very approximate, due to the large number of variables involved in the fitting procedure employed (Lankford and Does 2012), and the requirement of uniform transmit RF fields. For such reasons MWF mapping has not yet been used in cortical parcellation studies.
Measurement of magnetization transfer rate (MTR) has also been proposed as a method for myelin density quantification (e.g. Vavasour et al. 1998; Alexander et al. 2011). This method (Wolff 1989) relies on the fact that the spin–lattice relaxation of longitudinal magnetization in tissue arises largely from transfer of the spin magnetization of protons on free water molecules to protons bound to large relatively immobile biological molecules, mostly embedded in membranes. Transfer of spin magnetization in the opposite direction also occurs, from protons bound to the large molecules to the free water protons. This can be enhanced by applying an RF magnetic field at a frequency offset from the free water proton Larmor frequency, which excites the protons of hydrogen atoms attached to large molecules, having a broad resonance spectrum. The coupling of this spin magnetization to the free water protons accelerates their relaxation, and transfer of the magnetization saturates them, reducing the net magnetization. These effects on T1 and spin magnetization M0 thus provide an indication of the local molecular environment. In much of brain tissue, especially white matter, myelin dominates the local molecular environment, and so it has been considered that MTR also provides a map of brain myelin (e.g. Gringel et al. 2009; Helms et al. 2010). However, while there is a rough concordance between maps generated using MWF and MTR methods, they do not match in detail (Vavasour et al. 1998), giving rise to a concern that neither of these methods attain this goal. MTR imaging also tends to provide a relatively low SNR per unit time, because it compares the signal from images with rather similar intensity, with and without a magnetization saturation pulse.
Given the ubiquity and concentration of myelin in brain tissue, it is expected to affect MR image contrast in many ways. Table 7.1 (Siegel et al. 1999) provides the dry weight composition for several important constituents of brain tissue.
Table 7.1
Concentrations of lipids in gray matter, white matter, and myelin of human brains (Reprinted with permission from Siegel et al. 1999). Protein and lipid figures in percent dry weight; all others in percent total lipid weight
Myelin | White matter | ||||||
|---|---|---|---|---|---|---|---|
Substance | Human | Bovine | Rat | Human | Bovine | Gray matter (human) | Whole brain (rat) |
Protein | 30.0 | 24.7 | 29.5 | 39.0 | 39.5 | 55.3 | 56.9 |
Lipid | 70.0 | 75.3 | 70.5 | 54.9 | 55.0 | 32.7 | 37.0 |
Cholesterol | 27.7 | 28.1 | 27.3 | 27.5 | 23.6 | 22.0 | 23.0 |
Cerebroside | 22.7 | 24.0 | 23.7 | 19.8 | 22.5 | 5.4 | 14.6 |
Sulfatide | 3.8 | 3.6 | 7.1 | 5.4 | 5.0 | 1.7 | 4.8 |
Total galactolipid | 27.5 | 29.3 | 31.5 | 26.4 | 28.6 | 7.3 | 21.3 |
Ethanolamine phosphatides | 15.6 | 17.4 | 16.7 | 14.9 | 13.6 | 22.7 | 19.8 |
Lecithin | 11.2 | 10.9 | 11.3 | 12.8 | 12.9 | 26.7 | 22.0 |
Sphingomyelin | 7.9 | 7.1 | 3.2 | 7.7 | 6.7 | 6.9 | 3.8 |
Phosphatidylserine | 4.8 | 6.5 | 7.0 | 7.9 | 11.4 | 8.7 | 7.2 |
Phosphatidylinositol | 0.6 | 0.8 | 1.2 | 0.9 | 0.9 | 2.7 | 2.4 |
Plasmalogens | 12.3 | 14.1 | 14.1 | 11.2 | 12.2 | 8.8 | 11.6 |
Total phospholipid | 43.1 | 43.0 | 44.0 | 45.9 | 46.3 | 69.5 | 57.6 |
It is clear from this table that myelin concentration levels are high enough to affect proton density contrast. Myelin also has a strong effect on the transverse relaxation times T2 and T2*. T2 is primarily affected because the water between the myelin layers wrapping axons has a particularly short T2, as described above. The T2 of the non-myelin water in white matter is surprisingly similar to T2 in grey matter (Whittall et al. 1997).
It is still poorly recognized how little T2 contrast there really is in the brain, outside of deep brain structures containing iron (see below). Because its volume fraction is small (less than 15 %), and its T2 is short ( < 15 ms), myelin water contributes little to the value of T2 measured conventionally by comparing the signal at two echo times, or by measuring with the Carr-Purcell sequence. Instead, so-called “T2-weighted images” provided by MRI sequences such as turbo-spin-echo (TSE) typically gain much of their grey-white matter contrast from magnetization transfer (see Thomas et al. 2004; Turner et al. 2008), differences in T1, or proton density variations. Optimal contrast-to-noise for TSE sequences, which typically have echo trains extending much longer than T2, is obtained at the shortest possible echo times, which would not be the case if T2 difference were the major contrast mechanism. When T1 was measured using an inversion-recovery prepared turbo-spin-echo sequence, it was found that MTC effects reduced its value by about 30 % (Turner et al. 2008). It has been historically unusual to create genuine maps of either the short or long T2 component of the bi-exponential transverse relaxation decay curve. A very careful study was performed by Oros-Peusquens et al. (2008). Her quantitative maps of T2 in normal brain show very little overall contrast between grey and white matter, with a large overlap in their T2 distributions. See also Fig. 7.2, which shows a quantitative map of T2 in normal brain obtained at the Leipzig MPI at 7 T. The futility of attempting cortical parcellation using T2 maps alone is underscored by West et al. (2012) who show the enormous overlap of the T2 distributions in WM and GM.
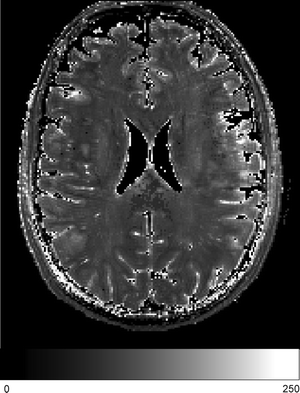

Fig. 7.2
Quantitative map of T2, measured at 7 T in an axial slice through normal human brain. Spatial resolution was 1 mm isotropic. Data obtained at five echo times were fitted to a single exponential decay curve. Grey scale gives T2 in milliseconds. Note the minimal grey-white contrast (Courtesy of Zaheer Abbas)
In practice, when magnetization transfer effects are carefully avoided, the grey-white contrast obtained by “T2-weighted sequences”, such as TSE, is dominated by variations in T1 and MTC at echo times longer than 100 ms. The main reason for this is that the echo train generated by the repeated radiofrequency refocussing pulses of the TSE sequence consists largely of stimulated echoes, for echo times longer than about 100 ms. Stimulated echoes decay with a time constant of T1, which is much longer than T2 in tissue, and thus the signal in these later echoes is heavily T1 weighted. Even with echo times as short as 26 ms (e.g. Trampel et al. 2011) TSE contrast is clearly dominated by magnetization transfer effects.
T2* weighted images and T2* maps of the brain, on the other hand, show highly significant signal variations, both between grey and white matter and within each of these tissues. This reflects the presence of myelin, but also another very important source of MRI contrast in brain, iron (Drayer et al. 1986). It is now becoming clear that the main origin of T2* contrast is that the magnetic susceptibility of lipid molecules, and of commonly found iron-containing molecules, particularly ferritin, is different from that of water. (Duyn 2011; Lee et al. 2011) When the tissue experiences a static magnetic field, this difference causes microscopic field gradients to arise in water associated with such molecules. This is especially the case in WM, with a high density of lipid membranes forming myelin sheaths around the axons, and in deep brain nuclei, such as the substantia nigra, with high densities of iron in the form of ferretin and neuromelanin. The net result of the dephasing of the spin magnetization caused by these magnetic field gradients is a reduction in T2*. This susceptibility difference also partly explains the strong phase contrast between GM and WM (Li et al. 2011). T2* weighted images and phase images of the brain can be obtained with very high spatial resolution. Cortex often contains a significant quantity of myelinated axons (the basis of myeloarchitecture, indeed), and iron deposits are often associated with such cortical myelin (Fukunaga et al. 2010) (see Fig. 7.3).
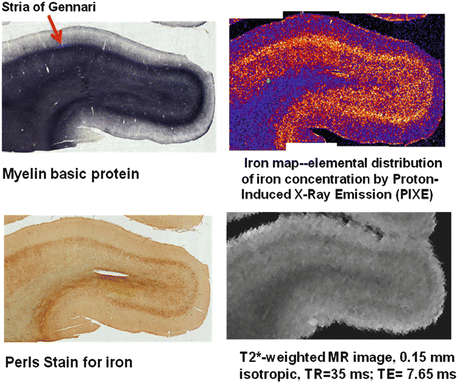

Fig. 7.3
Image data from a block of human cadaver brain tissue containing primary visual cortex, V1 (Courtesy of Carsten Stüber, Leipzig). The Stria of Gennari is prominent in this block. Ex-vivo MRI scanning was performed in Leipzig at 7 T (Siemens Magnetom). The brain block was then sliced. Adjacent slices were given various histological stains. One slice was analyzed using Proton-Induced X-ray Emission provided by a proton beam accelerator (Lipsion) to map elementary iron content within the slice. Note that cortical myelin is much richer in iron than white matter myelin, but both iron and myelin contribute to T2* contrast
Thus T2*-weighted images show excellent detail in cortical grey matter (Duyn 2011) (see Fig. 7.2), which has been shown to depend on the angle between B0 and the local axonal orientation (Cohen-Adad et al. 2012) (Fig. 7.4).
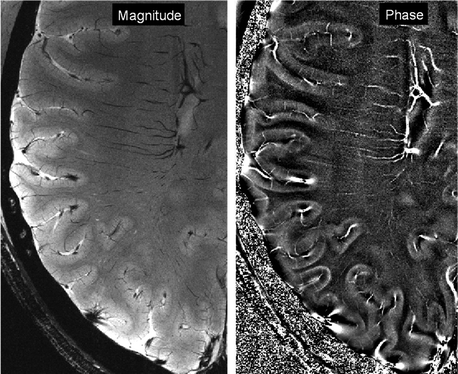

Fig. 7.4
Axial images of normal human brain acquired on a Siemens Magnetom 7 T scanner at Leipzig by Andreas Schaefer. FLASH sequence. In-plane resolution 0.23 × 0.23 mm2. Slice thickness 1 mm. 24-channel NOVA RF coil
But probably the most striking impact of myelin on MR images lies in its effect on T1. For precisely the same reason that it affects MTR, the presence of myelin substantially accelerates T1 relaxation, by the transfer of magnetization from slowly relaxing free water protons to rapidly relaxing protons bound to large molecules in the cell membranes. Those specific large molecules that are particularly responsible for this powerful relaxation mechanism have been discovered by careful study of model membrane systems comprised of phospholipids and water (Koenig 1991; Kucharczyk et al. 1994). When either of two important components of typical cell membranes, cholesterol and cerebroside, were added in biologically typical concentrations, they were found to dramatically shorten T1. These molecules have hydrophilic hydrogen-containing head groups that sit at the membrane-water interface and provide a conduit for the transfer of magnetization from the free water protons to the bulk of the membrane.
Thus so-called “T1-weighted” images have come to be regarded as the standard for depicting grey-white contrast in human brain, and used in numerous applications which require image segmentation of grey and white matter, such as voxel-based morphometry (Ashburner and Friston 2000). Generally low flip-angle gradient-echo (FLASH) sequences (Frahm et al. 1986) are used to provide such images. Here the contrast arises because white matter protons recover faster than grey matter protons to alignment with the static field, providing a larger equilibrium signal. The contrast can be further enhanced using a preparatory inversion pulse, such that the grey matter signal is close to being nulled at the time of image data acquisition, at which time the white matter signal has already partially recovered. The resultant sequence (Mugler and Brookeman 1990) is named MP-RAGE (magnetization-prepared rapid-acquisition gradient echo). However, it should be pointed out that neither FLASH nor MP-RAGE sequences actually produce maps of the value of T1. They have higher intensity where T1 is shorter, and their intensity is also weighted by proton density and T2* values.
In order to obtain quantitative maps of T1 and thus to provide a better guide to tissue myelin, each of these sequences must be modified. Adding a second excitation pulse with a different flip angle to the FLASH sequence (Preibisch and Deichmann 2009), or sampling the MP-RAGE signal at two different inversion times, allows the longitudinal relaxation time T1 to be mapped with reasonable precision. The second of these sequences is known as MP2RAGE (Marques et al. 2010). Maps of T1 may also be generated by acquiring a series of inversion-recovery T1-weighted images, and fitting the inversion time dependence in each voxel to an exponential function. Importantly, these maps show little effect of the inhomogeneity of image intensity (bias-field) normally caused by spatial variations in RF excitation and reception.
In conclusion, comparing MRI methods for detecting the presence of myelin, it can be hypothesized that maps of relaxation rate R1 ( = 1/T1) and MTR maps provide essentially identical information, proportional to the area of membranes in good contact with the free water pool (i.e. the outer and inner circumferences of myelinated axons), while MWF maps provide a good quantification of total myelin density. This concept merits much deeper exploration, including quantitative chemical assays of myelin density, in future research.
7.3 In-Vivo MRI Microscopy: Issues of Signal, Sequence and Field Strength
Whatever the source of contrast and the structures to be investigated, the most important consideration for useful cortical mapping is to have a sufficiently large contrast to noise ratio (CNR). Although fundamental limits exist, related to the density of water protons in brain tissue and the fact that MRI is performed at body temperature, achievable CNR can be improved by maximizing raw NMR signal, minimizing noise, optimizing the MRI sequence strategy, maximizing acquisition time, minimizing effects of brain motion, and optimizing the static magnetic field strength.
7.3.1 Signal
For a given water proton density, flip angle, static magnetic field uniformity, sampling rate, and relaxation times, the NMR signal depends linearly on the voxel volume, and the RF receiver coil sensitivity. Intracortical detail exists at a scale of less than 500 μm, and the cortical surface is always multiply curved. Thus cubic voxels of less than 500 μm are essential. At this resolution, using a very high performance MR sequence, Turner et al. (2008) showed that at least 30 min of scan time was required at 3 T to give enough CNR to detect the Stria of Gennari reliably in multiple slices with resolution 0.4 × 0.4 × 0.5 mm3, with the 12-channel RF receiver coil then available.
The receiver coil sensitivity itself depends largely on how close the coil elements are to the tissue to be imaged. Thus, other things being equal, to obtain good signal from an entire head it is desirable to have a large number of small coil elements, mounted on a coil former only just large enough to accommodate the head. These are configured in the form of a “phased array” (Roemer et al. 1990; Wiggins et al. 2009). With a 64 channel array an improvement in SNR of a factor about 2–3 is claimed in cortical areas relatively close to the skull, which should shorten the acquisition time for the isotropic detection of the Stria of Gennari at 3 T, as just described (Turner et al. 2008) to about 8 min.
7.3.2 Noise
Random noise in MRI is primarily white noise, arising from thermal fluctuations in the object to be imaged, in the conductive elements of the coil itself, and in the receiver preamplifiers. The present generation of receiver preamplifiers adds well below 1 dB of noise to the signal. Thermal noise can be reduced by making the receiver bandwidth as small as possible, since the noise is proportional to the square root of the bandwidth. There are two limits to this minimization. The first is that the NMR signal decays over a time of about 100 ms, so there is never any point in having a receiver bandwidth less than 10 Hz. Secondly, and more important for imaging, the acquisition readout window for a given phase-encoded echo is never greater than 100 ms, and within this window up to 512 data points must be obtained to sample the signal which has been spatially encoded by the read gradient. This means that the acquisition bandwidth is at least a few kilohertz. This has the advantage, for typical multi-pulse imaging, that the imaging magnetic field gradients are much larger than the endogenous field gradients produced by inhomogeneities of magnetic susceptibility within the head, so there is little spatial distortion in the final images.
7.3.3 MRI Sequence Choice
MR microscopy has been performed for several decades (reviewed in Blackband et al. 1999). With a short echo-time spin echo sequence, a field strength of 14 T, and an hour of scan time, spatial resolution of 1 μm in-plane was achieved ex vivo (Lee et al. 2001). Fresh interest in MRI neuroanatomy was generated by the publication by Fatterpekar et al. (2002) of high resolution images of cadaver brain at 9.4 T with a spatial resolution of 78 × 78 × 500 μm3. These images required 14 h of scan time. No attempt was apparently made to maximise the CNR per unit time, and the authors fail to state what type of sequence they used. From the contrast that they found, it is fair to assume that they used a single-slice gradient-echo sequence, with a stated echo time of 45 ms and a very long TR of 2.4 s. Such a sequence provides good grey-white matter contrast due to the shorter T2* in white matter, and good intracortical contrast due both to myelinated cortical layers and the associated iron deposition. However, owing to the lack of technical detail it is quite impossible to deduce a quantitative relationship between the MR contrast and the optical density of the corresponding stained sections which they show.
Fortunately, MRI physics is a science, and not a black art. It is possible to relate measured image intensity to the basic parameters of the Bloch equation, and in the present context it is highly desirable to do so (see also Fischl et al. 2004). The duration of an MRI scan in vivo is basically limited by the capacity of a human volunteer subject to remain immobile for an extended period of time. Thus the ideal MRI sequence for in-vivo cortical parcellation makes maximum use over time of the available spin magnetization, with minimum time spent preparing the spin magnetization, when data are not acquired. In addition, the highest possible contrast between grey matter and white matter is desired. Further requirements are that the image intensity should simply reflect an NMR parameter that can be used as an index of myelination, and that the RF power deposition should be the minimum required to generate sufficient contrast. This final consideration is associated with the need to use high magnetic field strength (see next section), and the fact that RF power deposition (SAR, specific absorption rate) increases as the square of the field strength.
It is an unfortunate historical fact that so far there has been no systematic investigation as to which sequence delivers the closest approximation to these requirements. The two main classes of sequence are (i) gradient echo sequences, lacking refocusing pulses, in which low flip angle pulses are repeated at short intervals, each followed with read gradients applied while data are acquired; and (ii) spin echo sequences, which typically use higher flip angle excitation pulses, with refocusing pulses to recover the spin magnetization dephased by local magnetic field gradients.
(i)
Gradient echo sequences. Here the three main contenders for high SNR and good grey matter/white matter contrast are the MP-RAGE sequence (Mugler), the modified version using a second inversion pulse, MP2-RAGE (Marques) and the Modified Driven Equilibrium Fourier Transform (MDEFT) sequence, optimized by Deichmann (2006). Acquisition parameters for the MP-RAGE sequence have recently been optimized by Bock et al. (2013), giving a substantial improvement in CNR per unit time.
(ii)
Spin echo sequences. Until the invention by Hennig et al. (1986) of the RARE sequence, gradient echo sequences (which include SSFP, FISP, PSIF, etc.) made more efficient use of the steady-state magnetization that was available, but the string of refocusing pulses following a single excitation pulse that form the essence of RARE (turbo-spin-echo (TSE), fast spin-echo, etc.) greatly increased the efficiency of this sequence. They also greatly increase the SAR, making standard TSE sequences difficult to use at field strength above 3 T.
Fortunately an alternative sequence to TSE exists, which gives nearly identical contrast, somewhat higher signal to noise per unit time, and has a much smaller RF power deposition. This is the GRASE sequence (GRAdient echo and Spin Echo) first described by Oshio and Feinberg (1991). This sequence is similar to TSE, in that it uses a train of refocussing 180° RF pulses, but most of these pulses are replaced by alternating gradients, as in echo-planar imaging, so that the signal forms a train of gradient echoes, which are periodically refocused by the 180° pulses. As with TSE, this echo train decays with a time constant closer to T1, which enables a long acquisition window, but the radiofrequency power deposition is drastically reduced. Trampel et al. (2010) has shown that for the same effective echo time, voxel size and TR time, the TSE and GRASE images of a brain are almost identical, so that image subtraction reveals only a few edges, and mostly noise. The GRASE sequence has great promise for ultra-high resolution scanning of human brain at very high field.
7.3.4 Maximizing Acquisition Time
Signal to noise can obviously be increased by longer averaging of image data. However, SNR improves only with the square root of the imaging time, so that scanning for an hour gives only double the SNR obtained in 15 min. And this apparent gain can easily be cancelled out by the increasing likelihood with scan duration that the subject will move significantly. One solution (Turner et al. 2008) is to average a number of separate scans after they have been co-registered with each other. Keeping one’s head still for 20 min is considerably easier than trying to remain immobile for an hour or more. In this approach, it is advisable to average in the image domain, but to retain the full complex data, so that the real and imaginary components of the image are separately averaged, and then recombined to form a magnitude image (Oros-Peusquens et al. 2010). In this way, data from as many as ten imaging sessions with a given human subject can be combined with very little loss of spatial resolution.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree








