Methods in Psychiatric Genetics
A scientific revolution has occurred in the field of genetics with the advent of molecular biological techniques. Using these techniques, genes influencing risk for many neuropsychiatric diseases have been identified: initially Mendelian single gene conditions such as Huntington disease were resolved; in the last few years complex genetic conditions such as alcohol dependence have yielded specific genes. Some of this work has been facilitated by the study of endophenotypes or biologic vulnerability markers.
Three types of population genetic studies—twin, family, and adoption studies—are conducted to ascertain whether a particular human phenomenon is substantially genetically influenced.
Twin studies are based on the fact that monozygotic (MZ) or identical twins represent a natural experiment in which two individuals have exactly the same DNA sequence for each of their genes. This is in contrast to dizygotic (DZ) or fraternal twins who share 50% of their DNA sequences and are no more genetically similar than any pair of siblings. A phenomenon that is influenced by genetic factors should be more “concordant” (similar) in MZ twins compared to DZ twins.
Family studies can answer three critical questions concerning the inheritance of a disorder:
- Are relatives of an affected subject at increased risk for the disorder compared to relatives of control subjects?
- What other disorders may share a common genetic vulnerability with the phenomenon in question?
- Can a specific mode of inheritance be discerned?
A family study typically begins with a proband or initially ascertained patient, whose relatives are then studied.
Adoption studies: In adoption studies the risk for the disorder may be evaluated in four groups of relatives: the adoptive and biological relatives of affected adoptees and the adoptive and biological relatives of control adoptees. If the disorder is heritable, one should find an increased risk among the biological relatives of affected subjects, compared to the other three groups of relatives. One can also compare risk for illness in adopted-away children of ill parents versus adopted-away children of well parents.
Segregation analysis may be used to determine whether the pattern of illness in families is consistent with a specific mode of transmission. This is most useful for conditions in which a single gene accounts for a substantial portion of the variance. Most major psychiatric disorders do not appear to fall in this category.
Some of the complexities of major psychiatric disorders are as follows:
- Variable penetrance (some individuals with a genetic predisposition will not manifest the disease)
- Phenocopies (individuals without a genetic predisposition who manifest the symptoms of the disease)
- Genetic heterogeneity (more than one type of genetic cause can produce the same syndrome)
- The diagnostic boundaries of a syndrome may be uncertain.
- Pleiotropy (one gene may be expressed in different ways in different persons).
At any genetic locus, each individual carries two copies (alleles) of the DNA sequence that defines that locus. One of these alleles is inherited from the mother and the other from the father. These alleles will be transmitted with equal probability (i.e., 1/2), one of the two alleles to each offspring. If two genetic loci are “close” to each other on a chromosome, their alleles tend to be inherited together (not independently) and they are known as “linked” loci. During meiosis, crossing over (also known as recombination) can occur between homologous chromosomes, thus accounting for the observation that alleles at linked loci are not always inherited together.
The rate at which crossing over occurs between two linked loci is directly proportional to the distance on the chromosome between them. In fact, the genetic distance between two linked loci is defined in terms of the percentage of recombination between the two loci (this value is known as theta). Loci that are “far” apart on a chromosome will have a 1/2 chance of being inherited together and they are not linked. Thus, the maximum value for theta is 0.5, while the minimum value is 0. Linkage analysis is a method for estimating theta for two or more loci.
The probability that two loci are linked is the probability that theta <0.5. The probability that the two loci are not linked is the probability that theta = 0.5. Thus, a LOD (logarithm of the odds ratio) score is defined as
Although it is possible to perform such calculations by hand (see Ott, 1985), LOD scores are usually calculated using computer programs, such as GENEHUNTER or Merlin. Since a LOD score is a log value, scores from different families can be summed. For complex conditions, collections of affected sib pairs may be studied rather than large families. A LOD score of 1.0 indicates that linkage is 10 times more likely than nonlinkage. For simple genetic conditions, a LOD score of three or greater is evidence for linkage, while a score of −2 or less is sufficient to exclude linkage for the sample studied. For disorders with more complex forms of inheritance (including most psychiatric disorders), a higher positive LOD score is required (3.6 for definite linkage and 2.2 for suggestive linkage).
In association studies one compares allele frequencies for a given locus in two populations, one of which is composed of unrelated individuals who have a disease, while the “control” population is usually composed of ethnically similar unrelated persons who do not have the disease. If a particular allele commonly predisposes individuals to the disease in question, then that allele should occur more frequently in the disease population, compared to the control population.
There are potential pitfalls to the case-control association approach. The locus chosen for study must predispose to illness. Thus, loci chosen for association studies are often known as candidate genes. If the locus does not predispose to illness, then the results of an association study should be negative. However, false positive results can occur if the two populations are not carefully matched for ethnic background. One alternative control group is the parents of affected individuals (the alleles not transmitted to the affected child compose the “control group”—this is known as the Transmission Disequilibrium Test or TDT).
Biochemical studies of individuals with psychiatric diseases are always confounded by the issue of disease effects: are biochemical differences between affected individuals and controls related to the cause of the disorder, or are they related to the effects of the disorder (or its treatment)? When investigating possible biochemical differences for a genetic disease, this difficult issue can be addressed by studying a group of individuals (usually adolescents or young adults) who are at high risk to develop the disorder under study (usually because they have parents and/or other relatives with the disorder). The high-risk group may then be followed over time to assess whether the biochemical abnormalities observed are truly predictive of the disease.
Simple sequence repeat (SSR) markers, also known as microsatellites represent a group of polymorphisms based upon a variable number of a repeated sequence. However, in the case of SSR markers, the repeated sequence consists of 2–5 nucleotides. The repeated sequence is often —(CA)n—, —(AG)n—, or —(AAAT)n—, although other SSR sequences have been described. The region containing the SSR is amplified by using a thermostable DNA polymerase in a polymerase chain reaction (PCR). Microsatellites are commonly used to test linkage.
A different class of DNA markers, single nucleotide polymorphisms (SNPs), are usually used to detect association. An SNP is variation at one base in a DNA sequence (e.g., compare GATACA with GATGCA, in which the fourth nucleotide can be either “A” or “G”). It is estimated that there are ≈3 million common SNPs in the human genome, evenly distributed across the 3 billion bases of the human genome.
SNP detection is now highly automated, permitting the determination of more than one million SNP genotypes on each DNA sample, via DNA “chips” (microscope slide shaped devices) to which are attached up to one million oligonucleotides (short fragments of DNA sequence ≈20–30 bases in length). These oligonucleotides each permit the determination of the presence of an allele at a single SNP. This advance now permits genome-wide association studies for complex disorders, in which the frequency of alleles at each of the one million SNPs are compared in large groups of cases and controls.
Genetic Studies of Specific Psychiatric Disorders
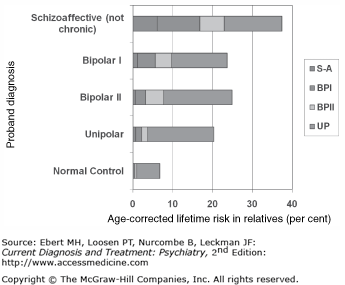
Family studies in affective disorder have continually demonstrated an aggregation of illness in relatives (Table 3–1). In a study at NIMH, 25% of the relatives of bipolar (BP) probands were found to have bipolar or unipolar (UP) illness, compared to 20% of relatives of UP probands and 7% of relatives of controls. In the same study, 40% of the relatives of schizoaffective probands demonstrated affective illness at some point in their lives. These data demonstrate increased risk in relatives of patients. They also show that the various forms of affective illness appear to be related in a hierarchical way: relatives of schizoaffective probands may have schizoaffective illness themselves, but are more likely to have BP or UP illness. Relatives of BP probands have either BP or (more likely) UP illness.
Age of onset may be useful in dividing affective illness into more genetically homogeneous subgroups. Early-onset probands have an increased morbid risk of illness in relatives in some data sets. Other subphenotypes, such as cycling frequency and comorbid anxiety disorders or substance use disorders, have also been studied.
A birth cohort effect was observed in several family studies, with an: an increasing incidence of affective illness among persons born more recently. The cohort effect was observed among relatives at risk to a greater degree than in the general population. The reasons for this increase in incidence are not yet clear.
Twin studies show consistent evidence for heritability. On the average, MZ twin pairs show concordance 65% of the time and DZ twin pairs 14% of the time.
Several adoption studies have been performed in the area of affective illness. The results have been generally consistent with genetic hypotheses.
Following are the types of affective disorders and other disorders that are genetically related:
Classic “manic–depressive illness” with severe mania, generally including episodes of major depression as well.
This disorder is genetically related to BPI and UP. There is some evidence in recent family studies for an excess of BPII illness in relatives of BPII probands. It has been demonstrated that BPII tends to be a stable lifetime diagnosis, that is, patients do not frequently convert to BPI.
Monozygotic Twins | Dizygotic Twins | |||
|---|---|---|---|---|
Reference | Concordant Pairs/Total Pairs | Concordance (%) | Concordant Pairs/Total Pairs | Concordance (%) |
Luxenberger (1930) | 3/4 | 75.0 | 0/13 | 0.0 |
Rosanoff et al. (1935) | 16/23 | 69.6 | 11/67 | 16.4 |
Slater (1953) | 4/7 | 57.1 | 4/17 | 23.5 |
Kallman (1954) | 25/27 | 92.6 | 13/55 | 23.6 |
Harvald and Hauge (1965) | 10/15 | 66.7 | 2/40 | 5.0 |
Allen et al. (1974) | 5/15 | 33.3 | 0/34 | 0.0 |
Bertelsen (1979) | 32/55 | 58.3 | 9/52 | 17.3 |
Totals | 95/146 | 65.0 | 39/278 | 14.0 |
Rapid-cycling BP illness has been the subject of great theoretical and clinical interest. A link with thyroid pathology has been proposed. Rapid-cycling appears to arise from factors which are separable from the genetic vulnerability to BP illness and which do not lead to aggregation within families. However “rapid switching” of mood, which is related, appears to be familial.
This entity includes BPI patients with no history of major depression. This group is not distinguishable from other BPI patients on the basis of family pattern of illness.
This condition of repetitive high and low mood swings, generally not requiring clinical attention, is probably genetically related to BP disorder.
A group of patients with intermittent psychosis during euthymia have an increase in affective illness and schizophrenia in relatives. This group may have the highest genetic load (total risk for affective or schizophrenic illness in relatives) of any diagnostic category. They may carry genes related to both BP illness and schizophrenia. Patients with chronic psychosis and superimposed episodes of mood disorder confer risk for both chronic psychosis and mood disorder to relatives but have less overall genetic load.
An overlap in linkage areas and vulnerability genes has been identified in recent years; some of this overlap may relate to genes involved in glutamate neurotransmission.
Family studies of anorexia and bulimia have generally found excess affective illness in relatives. Relatives of anorexics may have similar risk for affective disorders to that of relatives of BP probands.
Children with this disorder appear to have increased depression in their relatives. The opposite has not been demonstrated (BP/UP probands have not been reported to have increased risk of attention deficit disorder in their offspring).
There may be overlapping vulnerability traits. Alcoholism appears to be comorbid with UP and BP disorders (each appears to confer an increased risk for the other within individuals). There is some evidence that alcoholism with affective disorder may itself aggregate within families.
Linkage has been demonstrated on 4p, 6q, 8q, 13q, 18p, 18q, and 22q. Other areas are “close” to significant (e.g., 12q, 21q, and Xq).
A number of endophenotypic markers have been suggested, including:
- REM sleep induction by cholinergic drugs,
- white matter hyperintensities on MRI,
- amygdala activation on fMRI,
- hippocampal size,
- response to tryptophan depletion, and
- response to sleep deprivation.
Studies have begun on genome-wide gene expression in animal models of affective disorder, in brain samples from autopsy studies of patients with mood disorders, and in peripheral tissues such as blood. These studies should be helpful in identifying candidate genes for mood disorders.
More offspring of patients than controls have a diagnosed Axis I disorder. Offspring of BP parents may be more prone to respond to dysphoric feeling states with “disinhibitory” behavior.
Numerous candidate gene studies are now in the literature for BP illness. A few genes have emerged with replicated findings, or positive meta-analyses from multiple studies. We will feature these here.
This gene is one of the two implicated together in association studies on chromosome 13q. The gene G30 is a DNA sequence that is reverse-transcribed within G72. The association was first identified by Hattori et al. (2003) after work by Chumakov and colleagues in schizophrenia. It has been replicated by three other independent groups. The most recent work shows association not only with BP illness, but also with a subset of subjects who have schizophrenia with clear mood episodes. The function of G72 (also sometimes referred to as DAOA) may be to, oxidize serine, a potent activator of glutamate transmission via a modulatory site on the NMDA (n-methyl-d-aspartate) receptor. Inadequate DAOA function might be hypothesized to lead to problems in modulating the glutamate signal in areas of the brain such as the prefrontal cortex. Evidence from animal studies suggests that glutamate antagonists have antidepressant effects, and that depression is associated with inadequate modulation of glutamate neurotransmission. However a recent study suggests that the major role of G72 may be in maintaining neuronal structure.
This gene is a candidate based both on position (11p14, near reported linkage peaks in several family series) and function (as a neuronal growth factor, it is implicated in several recent theories of depression and BP mood disorder). BDNF has shown significant association with BP illness in three independent reports in family-based data, but not in several case-control series. Two reports have suggested association in child/adolescent-onset BP disorder, and two additional series show association in rapid-cycling BP patients. Several studies have shown that antidepressant administration is associated with increased central BDNF levels in experimental animals, and administration of BDNF itself has been associated with antidepressant-like activity. Depression has been postulated to be associated with decreased neurogenesis in the hippocampus, which is dependent on neurotrophic factors, including BDNF. Mood stabilizing medications used in BP illness are thought to have neuroprotective effects.
This gene on chromosome 1q was identified in a Scottish family with a genetic translocation and with multiple cases of psychiatric disorders, primarily schizophrenia. However DISC1 variants were associated with mood disorders in family members as well. Later studies in an independent series of BP patients in Scotland were positive for association as well. A study in Wales of schizoaffective patients showed a linkage peak in the same chromosomal location. This gene is expressed in multiple brain regions, including the hippocampus, where it is differentially expressed in neurons. It is associated with microtubules; in mice, disruption of DISC1 leads to abnormal neuronal migration in the developing cerebral cortex. DISC1 appears to interact with phosphodiesterase 4B, which may play a role in mood regulation.
These three genes have been shown in meta-analyses to be associated with BP disorder, even though no strong effects have been shown in any one study. The effect size for each appears to be in the range of 10–20% increase in risk. Each of these genes is associated with other behavioral phenotypes, and each has been reported to interact with environment to increase the risk of specific disorders (major depression, antisocial personality disorder, and schizophrenia respectively). Recent data in BP illness are more positive for 5HTT than for MAOA or COMT.
This gene on 12q24 was identified in a French–Canadian case-control series following linkage studies using large pedigrees from the same population. It is a calcium-stimulated ATPase. The data are suggestive, but await replication in an independent study.
This is the only candidate identified using animal model studies (a mouse model employing methamphetamine). The original gene expression studies were followed up by association studies in several samples as well as expression studies in human lymphoblasts. This gene participates in the down-regulation of G-protein coupled receptors.
Molecular genetic studies hold great promise in the future for families with affective disorder, particularly BP disorder. However genetic counseling currently is based on empirical risk figures.
The lifetime risk for severe (incapacitating) affective disorder is about 7%. Risk is increased to about 20% in first-degree relatives of UP patients, and 25% in first-degree relatives of BP. It appears to be 40% in relatives of schizoaffective patients. The risk to offspring of two affectively ill parents is in excess of 50%. Overall risk figures appear to be rising in recent years, but more so in relatives of patients than in the general population (keeping at about a 3:1 ratio). Average age of onset is about 20 for BP disorder and 25 for UP.
Twin studies tend to show heritability of drinking behavior and heritability of alcoholism. A Finnish twin study included interview data on 902 male twins between 28 and 37 years of age. Heritability was 0.39 (i.e., about 39% of the variance between members of a twin pair is due to genetic factors) for frequency of drinking and 0.36 for amount consumed per session. A second Finnish study involved several thousand pairs of twins in the state twin registry. Overall heritability for total alcohol consumption was 0.37 in males and 0.25 in females. A study in which 572 twin families from the Institute of Psychiatry register were examined found that additive genetic factors accounted for 37% of the variance in alcohol consumption among drinkers, when pedigree data are considered together with twin data and the effect of shared environment on twin concordance is accounted for. The critical data from these three large twin studies are strikingly similar, at least in males.
Twin studies of alcoholism itself have generally shown heritability. Kaij studied registration of twin subjects at the Swedish County Temperance Boards. Such registration implies that a complaint was made about a person’s behavior while drinking, either by the police or a third party. This would not generally include alcoholics who are socially isolated, though they might be significantly impaired. The registration information was followed up with personal interviews of probands and cotwins. In a total of 205 twin pairs, probandwise concordance was 54.2% in MZ’s and 31.5% in DZ’s (p <.01). Concordance rates in MZ’s increased with the severity of the disturbance. A reanalysis of these data shows heritability to vary from 0.42 to 0.98, the more serious forms of alcoholism being more heritable.
Kendler conducted a population-based study of female twin pairs from the Virginia twin registry. Personal interviews were completed on 1033 of 1176 pairs. MZ concordance varied from 26% to 47% (depending on whether a narrow or broad definition of alcoholism was used) while DZ concordance ranged from 12% to 32%. The calculated heritability was 50–61%. This suggests a substantial genetic influence for alcoholism in women.
Goodwin compared 55 adopted-away male children of an alcoholic parent with 78 adoptees without an alcoholic parent. The groups were matched by age, sex, and time of adoption. The principal finding was that 18% of the proband group were alcoholic compared with 5% of the controls (p <0.02). This study also compared adopted-away sons of alcoholics with sons of alcoholics raised by the alcoholic parent. There was no difference.
Bohman used state registers in Stockholm to study 2324 adoptees born in that city between 1930 and 1949. Male adoptees whose fathers abused alcohol (excluding those who were also sociopathic) were more likely to be alcoholic themselves (39.4% vs. 13.6%, p <0.01) compared with adoptees without an alcoholic (or sociopathic) father. Cloninger, Bohman, and Sigvardsson postulated a familial distinction of alcoholics: a milieu-limited (type I) and a male-limited (type II) group. Type I alcoholics usually have onset after age 25, manifest problems with loss of control, and have a great deal of guilt and fear about alcohol use. Type II alcoholics have onset before age 25, are unable to abstain from alcohol, and have fights and arrests when drinking, but less frequently show loss of control and guilt and fear about alcohol use. Cloninger reanalyzed the Stockholm Adoption data using these specified categories. He showed that type I alcoholics were significantly greater in prevalence only among those adoptees with both genetic and environmental risk factors (i.e., alcoholism in both biologic and adoptive parents). Type I was the most common type of alcoholism; however, it was present in 4.3% of controls with no risk factors. Type II alcoholism was present in only 1.9% of the controls but in 16.9–17.9% of adoptees with genetic risk factors. The presence or absence of environmental risk factors (alcoholism in adoptive parents) did not appear to make a difference.
Bohman extended this finding to women adoptees, identifying as particularly important the incidence of alcoholism in the biologic mothers of these adoptees.
There is a concentration of alcoholics in the families of alcoholic probands. Cotton (summarizing 39 studies on families of 6251 alcoholics and 4083 nonalcoholics) reports an overall prevalence of 27.0% alcoholism in fathers of alcoholics and of 4.9% in mothers; 30.8% of alcoholics had at least one alcoholic parent. The same preponderance of alcoholism was not seen in the parents of comparison groups of patients with other psychiatric disorders. The studies of nonpsychiatric controls reviewed in the same study show alcoholism rates of 5.2% in fathers and 1.2% in mothers. A recent report from the Collaborative Study of the Genetics of Alcoholism (COGA) shows significant coaggregation of drug dependence, mood disorders, and anxiety disorders as well as alcohol dependence in the relatives of persons with alcohol dependence.
Winokur reported an increased prevalence of depression in the female relatives of alcoholics, roughly comparable to the increased prevalence of alcoholism in male relatives. Some forms of illness may result from shared vulnerability factors. Recent studies suggest that comorbid disorders (including alcoholism and affective illness) themselves run in families.
Bohman and Cloninger observed that adopted-away daughters of type II (male-limited) alcoholics manifest no increase in alcoholism but do show an increase in somatization disorder.
It is not possible to conclude at this time that a single genetic predisposing factor is manifest as either alcoholism or sociopathy (antisocial personality disorder). However, some sociopathic alcoholics may transmit both alcoholism and sociopathy as part of the same syndrome.
Earls reported an increase in the Diagnostic and Statistical Manual of Mental Disorders, third edition (DSM-III) behavior disorder in general (attention deficit disorder with hyperactivity, oppositional disorder, and conduct disorder) in the offspring of alcoholic parents. The risk is greater for offspring of two alcoholic parents than for those of one alcoholic parent.
Several linkage studies have been completed in sizeable populations. Genes predisposing to alcohol dependence appear to be located on chromosomes 1, 2, 4, 7, and 16.
Variants in GABRA2 on chromosome 4p have been shown by Edenberg and colleagues in COGA to be associated with the power of beta oscillations in the EEG (which are inversely related to inhibitory neuronal activity in the cortex) and to alcohol dependence. This association has now been replicated by four other groups. GABRA2 appears to be particularly strongly related to problems with impulse control; the risk allele is also seen in adolescents with conduct disorder and in alcohol dependent persons who are drug dependent. Other GABA receptor genes such as GABRG3 may also be associated with alcohol dependence.
ADH (alcohol dehydrogenase) is the major metabolic enzyme for alcohol, catalyzing its breakdown into acetaldehyde, which is then further metabolized by aldehyde dehydrogenase (ALDH). Both ADH and ALDH have variants associated with the “flushing” reaction to alcohol (a feeling of warmth accompanied by reddening of the skin and sometimes nausea and tachycardia). These variants are most common in East Asian populations. They tend to protect against the development of alcohol dependence. In recent studies, single nucleotide polymorphisms in some of the ADH enzymes (genes for several isoenzymes of ADH are located on chromosome 4q) have been associated with alcohol dependence in Caucasian populations and in Native Americans. The strongest finding is in ADH4, which appears to be associated with the early onset of regular drinking.
The M2 muscarinic receptor gene on chromosome 7q was associated with alcohol dependence and major depression in the COGA study, an association that has been replicated. The association with depression recalls the cholinergic-adrenergic balance hypothesis of Janowsky and colleagues from the 1970s (in which a relative increase in central cholinergic activity is associated with depression and a relative increase in central adrenergic activity with mania).
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree