Fig. 1
Library enrichment for target-specific variants. The DNA encoding the initial capsid library (top left) is cloned into the vector and enters the cycle through which target-specific variants are enriched. At every new cycle, the complexity of the new enriched library decreases while the prevalence of target-specific variants increases
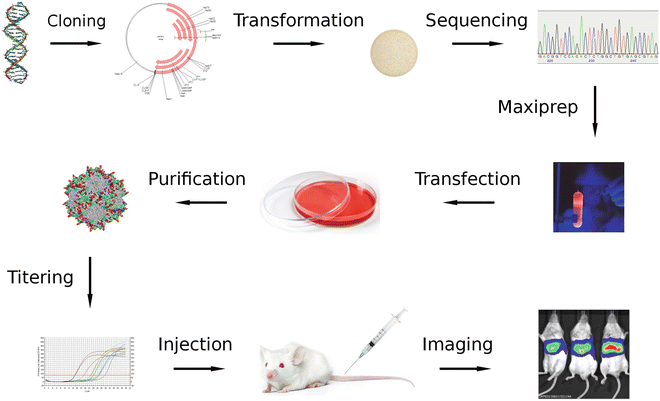
Fig. 2
Variant analysis and evaluation. Enriched capsid gene variants (top left) are cloned into a vector that does not contain ITRs. After transformation, clones are analyzed by sequencing to determine the most interesting variants, which capsid genes are then used to generate recombinant AAV that express luciferase. After injecting equal amounts in animals, specificities and transduction efficiencies of the different variants can be compared by monitoring luciferase activity using in vivo imaging
2 Materials
2.1 Plasmid Assembly
1.
pSubEagApa plasmid (see Note 1 ) that has been linearized with restriction enzymes EagI and ApaI and gel-purified.
2.
2.5× Isothermal DNA Assembly (IDA) buffer (see Note 2 ): for 1.44 ml, mix 664.62 μl H2O, 500 μl 1 M Tris–HCl pH 7.5, 10.38 μl 4.82 M magnesium chloride, 100 µl 10 mM dNTP, 50 μl 1 M Dithiothreitol, 625 μl 40 % PEG 8000, 50 μl 100 mM NAD. Store at −20 °C.
3.
1.33× IDA master mix (see Note 2 ): for 375 μl, mix 118.55 μl H2O, 200 μl 2.5× IDA buffer, 0.2 μl 10 U/μl T5 exonuclease (Epicentre, Madison, WI, USA), 6.25 μl Phusion 2 U/μl DNA polymerase (New England Biolabs, Ipswich, MA, USA), 50 μl 40 U/μl Taq ligase (New England Biolabs, Ipswich, MA, USA). Aliquot in PCR tubes at 7.5, 15, and 30 μl per tube, store at −20 °C.
4.
DNA Clean & Concentrator kit (Zymo Research, Irvine, CA, USA).
2.2 Transformation
1.
Electrocompetent E. coli: E. cloni 10G SUPREME (Lucige, Middleton, WI, USA) or equivalent (see Note 3 ).
2.
Electroporation cuvettes: 2 mm gap.
3.
Electroporation instrument: Bio-Rad Gene Pulser Xcell or equivalent.
4.
LB (Lysogeny Broth, also known as Luria Broth or Luria-Bertani medium): 10 g/L sodium chloride, 10 g/L tryptone, 5 g/L yeast extract. Sterilize by autoclaving.
5.
LB-Carbenicillin agar plates: 15 g/L agar in LB, 100 mg/L carbenicillin.
6.
LB with 100 mg/L carbenicillin.
2.3 Maxiprep
1.
Solution I: 50 mM Tris–HCl pH 7.5, 10 mM EDTA.
2.
Solution II: 0.2 N NaOH, 1 % SDS.
3.
Solution II: 3 M cesium chloride, 1 M potassium acetate, 0.67 M acetic acid (for 1 L: 505.08 g cesium chloride, 98.4 g potassium acetate, 38.46 ml glacial acetic acid).
4.
10 mg/ml RNase A solution.
5.
250 ml flat-bottom centrifuge tubes.
6.
Gauze sponges.
7.
Isopropanol.
8.
TE buffer: 10 mM Tris–HCl pH 8.0, 1 mM EDTA.
9.
Cesium chloride.
10.
10 mg/ml ethidium bromide solution.
11.
4.9 ml ultracentrifuge tubes and NVT90 rotor (Beckman, Indianapolis, IN, USA) or equivalent.
12.
18 G needles and 5 ml syringes.
13.
Isoamyl alcohol.
14.
Corex centrifuge tubes.
15.
Ethanol.
16.
Phenol-chloroform: phenol/chloroform/isoamyl alcohol solution (25:24:1).
17.
Chloroform.
18.
3 M sodium acetate, pH 5.2.
19.
75 % ethanol.
20.
SmaI restriction enzyme.
2.4 Transfection
1.
Human Embryonic Kidney (HEK) 293 cells (Sigma-Aldrich, St. Louis, MO, USA) grown in 150 mm tissue culture dishes.
2.
Culture medium: Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 5–10 % Fetal Bovine Serum (FBS) and Antibiotic-Antimycotic mix (final concentrations: 100 Units/ml each penicillin and streptomycin, 250 ng/ml Fungizone).
3.
2×HBS: 280 mM sodium chloride, 50 mM Hepes, 1.5 mM sodium phosphate dibasic. Adjust pH to 7.17 using 10 M sodium hydroxide (approximately 1.6 ml for 1 L). Sterilize by filtration through a 0.22 μm filter. Aliquot in 50 ml tubes and store at −20 °C.
4.
2.5 M calcium chloride. Sterilize using a 0.22 μm syringe filter. Store at −20 °C.
5.
pHelper plasmid (Agilent Technologies, Santa Clara, CA, USA).
6.
Standard cell culture equipment (CO2 incubator, sterile hood, centrifuge, water bath) and reagents (PBS, Trypsin).
7.
Lysis buffer: 150 mM sodium chloride, 50 mM Tris–HCl, pH 8.5.
2.5 Virus Purification
1.
0.22 μm vacuum filter.
2.
Tangential flow filtration system such as Minim II with a 100 kDa membrane such as 100 K Omega Centramate cassette (Pall Corporation, Port Washington, NY, USA).
3.
Benzonase (Sigma Aldrich, St. Louis, MO, USA) or equivalent nuclease.
4.
Saturated magnesium chloride solution: 4.82 M magnesium chloride. Store at room temperature.
5.
60 % Iodixanol solution: OptiPrep (Sigma-Aldrich, St. Louis, MO, USA).
6.
PBS-MK buffer: 1× phosphate- buffered saline (PBS), 1 mM magnesium chloride, 2.5 mM potassium chloride. Prepare as a 10× concentrated stock solution.
7.
40 % Iodixanol in PBS-MK.
8.
25 % Iodixanol in PBS-MK.
9.
15 % Iodixanol in PBS-MK and 1 M sodium chloride.
10.
Multichannel peristaltic pump such as Watson Marlow 205S or equivalent.
11.
29.9 ml ultracentrifuge tubes and 70 Ti rotor (Beckman or equivalent).
12.
18 G needles and 10 ml syringes.
13.
Lactated Ringer’s solution.
14.
Centrifugal quantitative concentrators: Apollo 20 ml, 150 kDa (Orbital Biosciences, Topsfield, MA, USA).
15.
0.22 μm sterile filters.
16.
Sterile low retention 1.5 ml microcentrifuge tubes.
2.6 Determination of Viral Titer
1.
DNase I and its 10× reaction buffer.
3.
DNA Clean & Concentrator kit (Zymo Research, Irvine, CA, USA).
4.
qPCR instrument, such as Bio-Rad MyiQ (Bio-Rad, Hercules, CA, USA).
5.
qPCR reagent, such as ABsolute Blue qPCR SYBR Green Fluorescein Mix (Thermo Fisher Scientific, Waltham, MA, USA).
6.
qPCR standard: linearized plasmid containing the target sequence, at known concentration, aliquoted in low retention tubes and stored at −20 °C.
7.
qPCR primers targeting AAV2 rep gene (to quantify capsid library titers): 5′-gcaagaccggatgttcaaat (forward) and 5′-cctcaaccacgtgatccttt (reverse).
8.
qPCR primers targeting the CBA promoter (to quantify capsid variant titers): 5′-tcccatagtaacgccaatagg (forward) and 5′-cttggcatatgatacacttgatg (reverse).
2.7 In Vivo Selection
1.
Young adult (6–10 weeks old) male mice.
2.
1 ml syringes with 25–28 Gauge needles.
3.
Induction chamber.
4.
Isoflurane.
5.
Scissors. Forceps.
6.
PBS.
8.
1.2 % sodium dodecyl sulfate (SDS) solution.
9.
Proteinase K (lyophilized).
10.
56 °C shaking incubator or water bath.
11.
10 mg/ml RNase A solution.
12.
0.45 μm syringe filters.
13.
Plasmid midiprep kit: EZNA Plasmid Midi kit (Omega Biotek, Norcross, GA, USA) or equivalent.
14.
DNA Clean & Concentrator kit (Zymo Research, Irvine, CA, USA).
2.8 Capsid Gene Amplification and Purification
1.
High fidelity DNA polymerase such as Q5 or Phusion (New England Biolabs, Ipswich, MA, USA).
2.
Standard PCR components and accessories: dNTP, PCR-grade water, PCR tubes, thermal cycler.
3.
PCR primers: 5′-ggatgggcgacagagtcatc (forward) and 5′-caagcaattacagattacgagtcagg (reverse).
4.
Standard DNA electrophoresis equipment: electrophoresis apparatus, power supply, agarose, TAE, buffer, loading dye, DNA ladder.
5.
Gel extraction kit (Omega Biotek, Norcross, GA, USA) or equivalent.
2.9 Variant Analysis
1.
pACG2-m56 plasmid (see Note 4 ) that has been linearized with restriction enzymes EagI and ApaI and gel-purified.
2.
Plasmid miniprep kit.
3.
PstI restriction enzyme.
4.
Sequencing primers 5′-gtctaggaactggcttcctg and 5′-ccattgaggtggtacttggtag.
2.10 In Vivo Evaluation
2.
Modified Dulbecco’s Phosphate Buffered Saline (mDPBS) without calcium chloride and magnesium chloride (Sigma-Aldrich, St. Louis, MO, USA).
3.
Luciferin solution: 15 mg/ml firefly luciferin sodium salt in mDPBS. Sterilize by filtering through a 0.22 μm syringe filter, aliquot in 2 ml tubes and store at −20 °C.
4.
In vivo imaging instrument such as IVIS Imaging System (Perkin-Elmer, Waltham, MA, USA).
3 Methods
3.1 Plasmid Assembly
See Note 6 for background information and explanations on why this particular method is used.
1.
Assemble reaction on ice by adding vector and insert (collection of capsid gene variants) DNA to one or more IDA master mix tubes (see Note 7 ) and adjust volume with water. See Table 1 for suggested amounts. Centrifuge briefly to collect reaction mixture to the bottom of the tubes.
Table 1
IDA reaction assembly and purification. Recommended parameters for IDA reaction assembly and purification depending on application, from initial library through enriched libraries and finally to variant analysis
Initial library | First enriched library | Second and subsequent enriched libraries | Variant analysis | |
|---|---|---|---|---|
Number and type of IDA master mix tubes | >10 × 30 μl | 1 × 30 μl | 1 × 15 μl | 1 × 7.5 μl |
Vector identity | pSubEagApa | pSubEagApa | pSubEagApa | pACG2-m56 |
Vector amount per tube | 200 ng | 200 ng | 100 ng | 25 ng |
Insert amount per tube | 150 ng | 150 ng | 75 ng | 19 ng |
Final reaction volume per tube | 40 μl | 40 μl | 20 μl | 10 μl |
Elution volume after DNA purification | 500 μl | 100 μl | 50 μl | 30 μl |
3.
Purify DNA from reactions using DNA Clean & Concentrator kit. Use at least 200 μl binding buffer. Elute with sterile water (not elution buffer), see Table 1 for recommended amount.
3.2 Transformation
1.
Place competent cells on ice (see Note 9 ). The total volume of competent cells, expressed in microliter, should be at least 4 times the amount of vector DNA used in plasmid assembly, expressed in ng.
2.
Place appropriate number of electroporation cuvettes on ice at least 15 min in advance.
3.
Add a volume of LB equal to 5 times the total volume of competent cells to a sterile flask and place on ice. Alternatively, aliquot the LB into several smaller flasks or tubes (the number of electroporation cuvettes should be a multiple of the number of flasks or tubes). Choose the size of the containers so that the LB does not fill more than 20 % of their total volume.
4.
Once the competent cells are thawed (if previously frozen), add purified DNA from Subheading 3.1 to the cells, mix gently, and aliquot into the electroporation cuvettes.
5.
6.
Incubate 1 h at 37 °C in a shaking incubator (220 rpm).
7.
Place an LB-carbenicillin agar plate under a sterile hood to warm up to room temperature and dry excess moisture.
8.
Transfer the transformation to a volume of LB-carbenicillin at least equal to 5 times the volume of the transformation and mix. If necessary, aliquot into several flasks so that each flask is filled at 20 % of its maximal volume.
9.
Spread 50 μl into the LB-carbenicillin agar plate.
10.
Incubate the agar plate at 37 °C overnight.
12.
Next morning, count colonies on the agar plate. Knowing the total volume of medium from which the aliquot was plated, calculate the complexity (extrapolated total number of colony forming units). Knowing the amount of vector DNA that was used, calculate the transformation efficiency (cfu/μg).
13.
If needed, Sanger or PacBio sequencing may be performed from colonies and from the liquid culture respectively.
3.3 Maxiprep
The following instructions assume an overnight culture volume of 1 L. Adjust the values according to the actual volume.
1.
Save 2 × 1 ml aliquots as glycerol stocks at −80 °C.
2.
Centrifuge at 5,000 × g for 15 min and discard supernatant.
3.
Completely resuspend pellet in 50 ml Solution I and 500 μl RNase A solution using a 10 ml pipette.
4.
Transfer to a 250 ml Beckman centrifuge tube.
5.
Add 50 ml Solution II and gently mix.
6.
Incubate on ice for 5 min.
7.
Add 50 ml Solution III and immediately mix, using the pipette in a stirring motion.
8.
Incubate on ice for 5 min.
9.
Centrifuge at 12,000 × g for 10 min.
10.




Transfer supernatant to a new 250 ml Beckman tube by filtering through a gauze sponge.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree






