Chapter 13 Genetics of Sleep in a Simple Model Organism
Drosophila
Abstract
In contrast to studies of naturally occurring genetic variation, one can attempt to induce mutations in animal models to test whether a given gene is important for sleep. One strategy to understand the molecular basis of complex behavior such as sleep is classic or forward genetics.1 Here a population of animals is randomly mutagenized using DNA alkylating agents such as ethyl methane sulfonate (EMS) or a mobile DNA transposable element. This mutagenized population is screened for a mutant phenotype of interest, such as altered sleep. Using molecular genetic approaches, one can then identify the mutant gene responsible for the mutant phenotype. Thus, forward genetics can be used to establish causal relationships between the function of individual genes and otherwise complex phenotypes. Forward genetics is unbiased and does not require any prior knowledge about the genetic basis of the phenotype of interest and is therefore an ideal approach for studying sleep. In contrast, reverse genetics starts with a disrupted gene in search of a phenotype. Nonetheless, the finding of a gene can provide insight into biochemical and cellular pathways that are important for sleep, perhaps even providing novel diagnostic tests or targets for drug development for sleep disorders.
Drosophila as a Model System for Genetics
Many of the model organisms in genetics, such as zebra-fish (Danio rerio) and the nematode Caenorhabditis elegans, have been adopted for sleep studies because they are highly suited to the forward genetics approach.2 Here I will focus on one of the premier model systems for genetics, the fruit fly Drosophila melanogaster (Fig. 13-1, Video 13-1![]() ). The fruit fly has been a workhorse for genetic studies since pioneering studies of Thomas Hunt Morgan in the early 20th century.3 A major advantage of Drosophila over many alternative model systems is the ability to grow and handle large numbers relatively easily and cheaply.1 A single female can produce hundreds of offspring. In addition, it has a short generation time, about 10 to 12 days from fertilized egg to fertile adult at room temperature. Because of these traits, Drosophila has been a model par excellence for high throughput screening of mutants with altered phenotypes. The facility of genetic mapping, gene disruption using transposable elements (mobile DNA), and the full genome sequence allows one to identify mutant genes responsible for mutant phenotypes.4 Remarkably (to some), the mammalian homologues of Drosophila genes have been found to function in a similar manner to their Drosophila counterparts. Indeed, entire signaling pathways are shared between Drosophila and their mammalian counterparts. For example, most human disease genes have clear fly homologues.5 Thus, genes identified in flies will likely serve comparable functions in more complex mammalian systems, including that of humans.
). The fruit fly has been a workhorse for genetic studies since pioneering studies of Thomas Hunt Morgan in the early 20th century.3 A major advantage of Drosophila over many alternative model systems is the ability to grow and handle large numbers relatively easily and cheaply.1 A single female can produce hundreds of offspring. In addition, it has a short generation time, about 10 to 12 days from fertilized egg to fertile adult at room temperature. Because of these traits, Drosophila has been a model par excellence for high throughput screening of mutants with altered phenotypes. The facility of genetic mapping, gene disruption using transposable elements (mobile DNA), and the full genome sequence allows one to identify mutant genes responsible for mutant phenotypes.4 Remarkably (to some), the mammalian homologues of Drosophila genes have been found to function in a similar manner to their Drosophila counterparts. Indeed, entire signaling pathways are shared between Drosophila and their mammalian counterparts. For example, most human disease genes have clear fly homologues.5 Thus, genes identified in flies will likely serve comparable functions in more complex mammalian systems, including that of humans.
The conservation between flies and mammals extends to the nervous system. Although flies have only about 1/1,000,000 the number of neurons as humans (about 105 versus 1011), the fly and human genomes are surprisingly similar in gene number (14,000 versus ~22,000), with differences largely due to gene duplication.6 In fact, the fly brain uses comparable neuronal machinery, including neurotransmitters, ion channels, receptors, and signal transduction pathways. Consequently, flies have been used as valuable nervous system models for olfaction,7 vision,8 hearing,9 sexual behavior,10 synaptic transmission, axon guidance,11 and learning and memory.12 Flies have also been exploited as models for numerous human diseases including diabetes,13 aging,14 pain,15 Alzheimer’s disease,11 Parkinson’s disease,11 epilepsy,16 and fragile X mental retardation.17 Finally, flies have been successfully used to study the response to clinically important drugs such as ethanol,18 cocaine,18 and general anesthetics.19 In many of these cases, genes identified in Drosophila serve similar functions in mammalian systems.
Drosophila as a Model for Studies of Sleep
Studies of Drosophila sleep have been predicated on a small but noteworthy literature examining sleeplike states in other invertebrate models such as mollusks20 and in insects such as cockroaches21 and honey bees.22,23 These classical descriptions of sleep behavior formed the basis for pursuing similar studies in Drosophila.
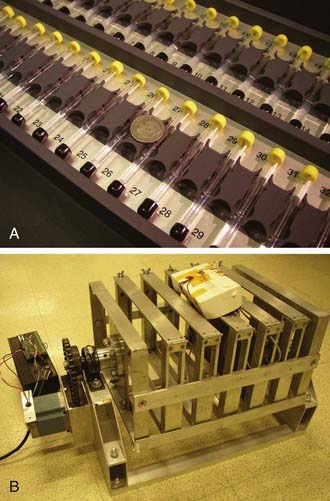
Sleep in the fruit fly is typically measured behaviorally using the Drosophila Activity Monitoring (DAM) System developed by Trikinetics (Waltham, MA; Fig. 13-2). Single flies are placed into a small transparent glass tube plugged on one end by agar food and the other end by a porous cap, allowing air passage. Each tube is placed into a monitor that contains a series of 32 infrared emitter-detector pair, one for each tube. An awake fly moves and back and forth in the tube, periodically breaking the infrared beam. Independent methods indicate a close correlation between infrared beam breaks and overall activity. A five-minute period of inactivity (no beam breaks) has been found to be a reliable indicator of sleep.
Given the remarkable similarity between Drosophila and mammals, it is not surprising that fruit flies exhibit many of the defining features of sleep. Flies exhibit extended periods of behavioral quiescence that can last for hours, and the majority of sleep typically occurs in bouts greater than 30 minutes.24 In addition, sleeping flies exhibit reduced responsiveness to sensory stimuli and exhibit homeostatic regulation.2,24–27 Video-based monitoring has also been coupled to measurements of beam breaks to provide higher spatial resolution analysis of fly sleep behavior.24,28
Drosophila sleep studies do not solely rely on measures of spontaneous movement; they also rely on responsiveness to sensory stimuli. Arousal threshold is assayed by application of a stimulus and measuring a behavioral response, typically induction of locomotor activity. During periods of extended immobility, flies are less likely to respond to a range of sensory stimuli including social, mechanical, vibratory, thermal, and visual.24,25,29,30 Although this responsiveness is typically measured behaviorally, it can also be uncoupled from movement using electrophysiologic measures.31 The typical fly demonstrates an increase in arousal threshold reaching a plateau after 5 minutes.25,30 The 5-minute criterion for sleep is in part based on this observation. Thus, one can distinguish quiet wakefulness from sleep by assessing arousal threshold.
Importantly, fly sleep is under homeostatic regulation: Flies deprived of sleep rebound the following day. Flies are typically deprived of sleep mechanically using automated devices or by tapping the flies by hand (see Fig. 13-2).24,25,30,32 The former must be programmed to vary the stimulus to avoid adaptation, and the latter allows one (with great patience and stamina) to deliver the stimulus selectively to sleeping flies. If a fly is deprived of sleep, it exhibits increases in sleep duration and intensity (the latter as measured by sleep bout length) the following day. Sleep rebound is not observed or is much less evident if similar deprivation protocols are applied to flies that are already awake, arguing strongly against nonspecific stress effects of mechanical disruption.24,25,30 Continuous sleep deprivation ultimately results in premature death in about 2 to 3 days32 as it does in some mammals.33 Unperturbed flies can live for 1 to 2 months. Thus, sleep is essential for life in the fly.
Although flies do not display the precise electroencephalographic signatures of mammalian sleep—the synchronous changes in neural activity seen as slow waves by electroencephalogram that are diagnostic of mammalian sleep have not been observed in Drosophila29—they do exhibit electrical correlates of sleep behavior providing behavior-independent state markers. In vivo electrical correlates in behaving flies have relied on the development of novel approaches to study the fly brain. In this approach, recording electrodes are inserted into the center of the Drosophila brain and into the optic lobes of a tethered fly (see Fig. 13-1).29 Local field potentials (LFPs) are measured, reflecting neural activity near the electrode. Leg movement in the tethered fly is simultaneously monitored. There is a general, but not perfect, correlation between spikelike potentials recorded from the central brain and waking movement. If flies are exposed to a rotating stripe, LFPs in the 20- to 30-Hz frequency are observed, reflecting attention to the stimulus, but these LFPs are reduced when the fly is asleep.31 In addition, periods of poor correlation between LFPs and movement are associated with increased arousal threshold and precede behavioral quiescence.31 These approaches clearly demonstrate that differences in arousal states can be characterized using electrophysiologic correlates as they are in more-complex organisms. The differences between fly and mammalian neuroanatomy could account for the differing electrical manifestation of sleep even if the underlying molecular and cellular mechanisms are similar. Studies of eye development provide some precedent for the idea that common genetic mechanisms can underlie anatomically distinct but functionally analogous structures.34
In addition to the core features of sleep, flies also display age-related changes in sleep architecture similar to those of aging mammals. Directly following emergence from the pupal case, young flies display elevated levels of sleep similar to their mammalian counterparts.25 With increasing age, sleep becomes more fragmented and less consolidated.35 In addition, drugs that increase oxidative stress can mimic these effects.35 Thus, the fruit fly has the potential to become a valuable model for the analysis of aging effects on sleep.
Mammalian sleep has been demonstrated to influence various aspects of memory consolidation.36 Similarly, flies also display sleep-loss–related deficits in learning and memory, using a number of different learning paradigms. For example, flies normally exhibit phototaxis but can be trained to avoid light using aversive stimuli. However, flies that have reduced sleep perform more poorly on this task.37 In courtship conditioning, male flies learn to stop courting females that have already mated. Under the appropriate conditions, males can remember this experience for over 24 hours; however, if flies are subjected to sleep deprivation after training (i.e., during the period of presumed memory consolidation), they fail to retain this memory.38 Finally, waking experience—in particular, social experience—can increase subsequent sleep amount, a process that might use dopamine and cyclic adenosine monophosphate (cAMP) pathways.38 Taken together, these data implicate a reciprocal relationship between sleep–wake regulation and synaptic plasticity memory in Drosophila, as is proposed in mammals.
Drosophila Circadian Behavior Reveals Conserved Mechanisms Between Flies and Humans
The best case for the argument that Drosophila genetics will illuminate the genetics of human sleep has emerged from studies of circadian behavior. As in many (but not all) organisms, sleep is under temporal control by a circadian clock in Drosophila.24,25 The first identified fly circadian mutants displayed short and long period rhythms in constant conditions and phase advanced and delayed activity in light–dark conditions, analogous to human advanced and delayed sleep phase syndromes.39,40 Cloning of the genes responsible for these fly phenotypes led to breakthroughs in our understanding of the core biochemical mechanisms of circadian timing. These studies also provide an experimental roadmap for elucidating basic mechanisms of sleep homeostasis. Although circadian clocks have often been viewed solely as timekeepers, both circadian genes and their accompanying neural circuits extensively regulate sleep–wake, perhaps independent of their timing functions (see later). Thus a deeper molecular understanding of the circadian system should provide insights into the control mechanisms for sleep.
Most aspects of the fly molecular clockwork are conserved with mammals, including humans (Table 13-1).41,42 Persons affected by familial advanced sleep phase syndrome (FASPS) exhibit an advanced phase of sleep–wake rhythms and shortened circadian period that is inherited in a mendelian dominant manner.40 Mutations in the human PER2 and CK1delta genes, orthologues of fly circadian genes period and doubletime, respectively, are responsible for this advanced sleep phase.43,44 These data argue that the basic architecture and core components of circadian clocks can be traced back to the shared ancestor of flies and humans hundreds of millions of years ago. Given the close association of circadian to sleep behavior, this suggests that the basic mechanisms of sleep homeostasis might also be conserved with flies.
Table 13-1 Drosophila Clock Genes and Their Highly Conserved Mammalian Homologues
| DROSOPHILA | MAMMALS |
|---|---|
| Period | Period1,2,3 |
| Timeless | Timeless |
| Clock | Clock, NPAS2 |
| Cycle | Bmal1 |
| Doubletime | CK1δ/ε |
| CK2 | CK2 |
| Cryptochrome | Cryptochrome 1,2 |
| Clockwork orange | Dec1, 2 |
| Slimb | β-TRCP |
Specific Neural Circuits are Important for Sleep–Wake Regulation
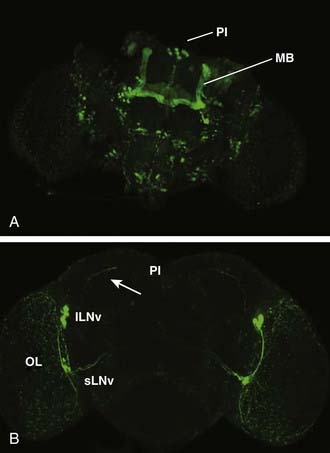
A theme of mammalian sleep studies is the notion that discrete neural circuits are important for initiating and maintaining sleep and wake states. Using gene-based tools to study neural circuit function in live animals, distinct neural circuits have been found that regulate sleep in Drosophila. Thus far, three anatomically defined loci have been implicated in sleep–wake regulation: the mushroom bodies (MBs), the pars intercerebralis, and the circadian pacemaker neurons: the large ventral lateral neurons (lLNv) (described earlier) (Fig. 13-3). In addition to these loci, additional circuits have been defined based on their transmitter identity (e.g., dopamine). These are discussed further later. To discover novel circuits involved in sleep regulation, an approach akin to forward genetics has been employed with the modification that, instead of screening for genes, circuits are screened in an unbiased manner for behavioral functions.
A cornerstone of this approach is the binary GAL4/UAS system.45,46 In one parental strain, the yeast transcription factor GAL4 is placed under the control of a tissue-specific promoter. In the second strain, the upstream activating sequence (UAS) bearing GAL4 binding sites is fused to an effector gene of interest. In the progeny of these two strains, the effector gene is expressed in the distribution specified by the tissue or circuit-specific promoter driving GAL4. Fortunately for Drosophila geneticists, there is a plethora of GAL4 lines available that provide a nearly limitless display of temporal and spatial expression patterns. In addition to the multitude of GAL4 lines, numerous UAS effector lines have been generated, including those that have been engineered to alter specific cellular properties such as membrane excitability or synaptic transmission. While these approaches are well developed in Drosophila, they have inspired even more sophisticated strategies in mammalian models.47
A number of cellular effectors have been successfully used for Drosophila sleep studies. One tool that has been used to conditionally block synaptic transmission is the UAS-driven shibirets1 (shits1) transgene.48 shi encodes for a multimeric GTPase homologous to mammalian dynamins. SHI is required for vesicle scission, a process that is required for synaptic vesicle recycling and thus the maintenance of fast synaptic transmission. UAS-driven expression of the shits1 allele in a wild-type neuron can block synaptic transmission at an elevated restrictive temperature (e.g., 29° C) but not at the permissive temperature (e.g., 21° C). Using the GAL4/UAS system and given the fact that flies are not homeotherms, one can specifically manipulate synaptic transmission in discrete neural circuits in a live behaving animal, using temperature acting as a remote control, and then assay the behavioral consequences of circuit modulation. Tools to manipulate cellular excitability have also been developed and applied. For example, a bacterial depolarization-activated sodium channel, NaChBac, has been used to increase cellular excitability.49 On the other hand, a non-inactivating mutant form of the voltage-gated Shaker potassium channel, termed electrical knockout (eko) has been used to silence neuronal activity.50
The use of these transgenic tools in combination with various GAL4 drivers led to the discovery of a sleep-regulatory role for the mushroom bodies, a bilateral neuropil well known for its role in learning and memory.12,51,52 To discover novel sleep-promoting circuits, a series of adult neural GAL4 lines were crossed with UAS-driven shits1.51 Their progeny were exposed to 12-hour cycles at restrictive (29° C) and permissive (21° C) temperatures, and the sleep behavior was examined. Reduced sleep at 29° C could be attributed to reduced synaptic transmission in the relevant neural circuits. Of nearly 100 lines tested, only a handful exhibited this reduced-sleep phenotype, suggesting that perturbation of neural function did not generally affect sleep behavior and that there are specific circuits in the fly devoted to promoting sleep. All of these lines displayed MB expression.
An independent method, chemical ablation of the MBs with hydroxyurea, was also used to assess MB sleep function. Hydroxyurea fed to larvae during the appropriate developmental time (first-instar) selectively ablates the neuroblasts that give rise to the large majority of the MBs.53 Like their shits1-expressing counterparts, flies fed hydroxyurea exhibited reduced sleep,51,52 and in both cases the reduced sleep was largely due to reduced sleep bout length—in other words, an inability to maintain sleep.51 In addition, flies with impaired or absent MBs exhibited a reduced lifespan consistent with a loss of restorative sleep.51 Thus, these flies are analogous to insomniac humans who are unable to maintain sleep and suffer adverse consequences as a result.
Although these observations suggest a sleep-promoting role for the MBs, other data using a different MB-GAL4 line suggest that the MBs might also promote wakefulness. These studies made use of a modified version of GAL4 called Gene Switch, in which the GAL4 is fused to the ligand-binding domain (LBD) of the progesterone receptor.54 In the absence of the ligand RU-486, the LBD retains GAL4 in the cytoplasm, rendering it inactive. In the presence of ligand, the LBD-GAL4 fusion is released to the nucleus, where the GAL4 binds its target UAS DNA sites and transcription can be initiated. RU-486 can be selectively delivered to adult flies by feeding to avoid any developmental effects of GAL4-driven gene expression. Using a version of Gene Switch that is driven by an MB-specific promoter,55 the silencing transgene eko resulted in increased sleep while expression of the activating NachBac transgene reduced sleep, suggesting that MB neuron activity increases wake.52 One possibility is that different subsets of MB neurons play opposing roles in sleep regulation. The finding that a part of the brain important for learning and memory is also important for sleep further supports the idea that sleep and memory consolidation are closely linked in the fly.
Flies that lack MBs demonstrate both spontaneous sleep (albeit reduced) and a robust homeostatic response, indicating that other brain loci promote sleep. One such locus is the pars intercerebralis (PI), a neuroendocrine cluster considered genetically analogous to the mammalian hypothalamus.56 Targeted decreases in epidermal growth factor (EGF) function in the PI result in reduced sleep.57 The PI may be a direct target of circadian pacemaker neurons. The dorsal projections of a subset of circadian pacemaker neurons, the small ventral lateral neurons (sLNv), terminate in close proximity to the PI neuron soma (see Fig. 13-3), and loss of the key peptide transmitter of these neurons, pigment dispersing factor (PDF), affects molecular circadian rhythms in PI neurons.58
In addition to sleep–wake circuits in the MBs and PI, the circadian pacemaker lLNv neurons promote wakefulness (see Fig. 13-3). Excitation of the lLNv using NaChBac or a novel tool, TrpA1, reduces sleep, especially at night.59–61 TrpA1 encodes for a cation channel that is activated at elevated temperatures, allowing conditional temperature-dependent regulation similar to the shits1 system.62 Selective ablation of the lLNv results in increased sleep.60 The vigilance affect is similar to that observed in animals in which the mammalian circadian pacemaker, the suprachiasmatic nucleus, is ablated.63 An important molecular effector of PDF+ pacemaker neuron functions in sleep is the transcription factor ATF-2, a member of the ATF/CREB (activating transcription factor/cAMP response element binding) family.64 The activity of these arousal-promoting neurons appears to be inhibited by gamma-aminobutyric acid (GABA),61 a relationship that is reminiscent of a similar organization of mammalian sleep circuits.65
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree