Fig. 8.1
The function of transcription factors in spinal motor neuron migration. Summary of expression of LIM homeodomain and Hox transcription factors in spinal motor neuron subpopulations (a). Functional evidence of the role of LIM homeodomain (b) and Hox transcription factors in spinal motor neuron soma localisation (c)
With the advent of these molecular markers it became possible to ask whether the transcription factors expressed by LMC, MMC and PG motor neurons themselves are contributing to the establishment of myotopic organisation. Thus, when the HB9 transcription factor was knocked out, its importance in the consolidation and maintenance of motor neuron identity became apparent, such that motor neurons lacking HB9 started to express interneuron markers and were found at aberrant positions within the spinal cord (Arber et al. 1999; Thaler et al. 1999). Manipulation of transcription factors that are confined to a spinal motor neuron column or division leads to a change in motor neuron position specification. Overexpression of the MMC identity marker Lim3 in LMC neurons causes them to adopt a cell body settling pattern like that of MMC neurons, in addition to inducing changes in molecular marker expression and axonal projection (Sharma et al. 2000). Similarly, overexpression of the lateral LMC marker Lim1 in medial LMC neurons causes their cell bodies to settle in lateral regions of the LMC (Kania and Jessell 2003), while loss of Lim1 from lateral LMC neurons causes them to adopt a more medial settling pattern (Palmesino et al. 2010). Ectopic expression of the medial LMC marker Isl1 in lateral LMC neurons causes them adopt a more medial position within the LMC (Kania and Jessell 2003). Together, these experiments argue that the LIM homeodomain transcription factor markers of spinal motor neurons are necessary and sufficient to specify a distinct settling pattern of spinal motor neuron subpopulations.
One of the earliest examples of transcription factors controlling motor neuron migration is that of Hoxb1, which is expressed in a population of motor neurons confined to the hindbrain rhombomere r4, and plays a role in specifying their migration pattern (Studer et al. 1996). In the spinal cord, rostrocaudally-restricted Hox transcription factors are also involved in the specification of motor neuron position. The observation that Hoxc6 is present in LMC motor neurons at brachial level and that Hoxc9 is expressed in PG motor neurons at thoracic levels, suggested that these transcription factors could be important in specifying PG versus LMC motor neuron fate, and thus, dorsomedial versus ventrolateral spinal cord position, respectively. Indeed Hoxc9 overexpression leads to a transformation of LMC neurons into PG neurons, including a shift in their position within the spinal cord. Likewise, Hoxc6 overexpression leads to the conversion of PG neurons into LMC neurons, along with a corresponding cell body position shift (Dasen et al. 2003).
Within a particular domain of the LMC, the rostrocaudal extent of motor pools also coincides with specific domains of Hox protein expression, implying that motor pool identity and therefore motor neuron migration patterns might also be controlled by this class of transcription factors. Hox5 and Hoxc8 expression forms a sharp boundary that coincides with the boundary of scapularis and pectoralis motor pools within the LMC. Overexpressing or inhibiting Hoxc8 shifts Hox5 expression and the boundary of these motor pools such that with the loss of Hoxc8 function, the caudal extent of the scapularis pool shrinks while the pectoralis pool expands more rostrally. Importantly, the mediolateral settling pattern of the ectopic domain of the pectoralis pool as assessed by the expression of its marker Pea3 transcription factor, appears more like the endogenous pectoralis pool and is markedly different from the scapularis pool (Dasen et al. 2005). Thus, not only do Hox genes control the identity of motor neurons and their cell body position pattern at the division level, i.e.: LMC versus MMC versus PG, but also can control it at the level of individual pools. This conclusion raises the question of whether the function of Hox genes at the level of entire motor neuron divisions can be disconnected from their apparently later action at the level of pools, or whether pool identity and soma settling pattern are defined simultaneously at divisional and pool levels. Perhaps the answer to this question might come from a better understanding of the soma position effector molecules whose expression is controlled by Hox genes.
One interesting idea that emerged from studying the development of motor neurons is that the transcription factor Foxp1 is required to mediate the function of essentially all Hox genes. Thus, in Foxp1 mutants, the above functions of Hox proteins in spinal motor neuron development are abrogated, leading to an interesting consequence on LMC neuron differentiation. In Foxp1 mutants, general spinal motor neuron identity is specified, along with a general localisation within the ventrolateral region of the spinal cord, and an axon projection through the ventral root. However, in such mutants, none of the above molecular markers of LMC neuron subpopulations are expressed, leading to the conclusion that Foxp1 removal results in the reversion of limb level LMC motor neurons to a more evolutionary primitive state in which they exist as a homogenous ensemble, undivided into subpopulations such as motor pools. Retrograde labelling of these motor neurons from a particular limb muscle reveals a disruption of their myotopic organisation, such that while motor neurons apparently innervating the same muscle are still located within the approximate position of the LMC, they are no longer located within the same cluster or pool in the ventral spinal cord (Dasen et al. 2008; Rousso et al. 2008). Of course, in the absence of any pool markers, it is difficult to conclude that these motor neurons are indeed part of a molecularly-defined motor pool in Foxp1 mutants. An alternative possibility is that motor neurons from a particular pool are still clustered, although their axonal projections in the limb are randomised. The resolution between these possibilities will develop as new Foxp1-independent molecular pool markers are uncovered.
The above experiments involve transcription factors that appear to be master regulators of spinal motor neuron identity at the level of divisions or at the level of pools. In addition to controlling cell body position, they sit at the top of a developmental hierarchy, controlling other aspects of motor neuron identity such as axonal projection. Indeed, gain and loss of function experiments of Foxp1, Lim1, Isl1 and HB9 (see above) all demonstrate that concomitant with inducing cell body position change, these manipulations also lead to a change of axon trajectory, but without any evidence of cross-talk between the soma migration and axon trajectory selection. In contrast, ETS transcription factors appear to control cell body position of spinal motor neurons in concert with axon trajectory selection. Initial experiments identifying these proteins as markers of specific motor pools demonstrate that their expression is dependent on signals from the periphery, such that limb bud ablation results in the extinction of expression of these transcription factors (Lin et al. 1998). Further experiments demonstrate that the neurotrophic factor GDNF, expressed in the limb, is actively signalling the maintenance of expression of the Pea3 ETS transcription factor, and that this maintenance is important for specifying the particular migration and dendritic arborisation pattern of the LMC motor neurons that express it (Livet et al. 2002; Haase et al. 2002; Vrieseling and Arber 2006). Thus, the ETS transcription factor Pea3 links axon growth into the limb to soma localisation, and therefore its action is mechanistically different from that of LIM homeodomain transcription factors whose expression does not appear to be induced by extrinsic signals dependent on axonal trajectory (Kania et al. 2000).
The converse of this idea is the dependence of lateral versus medial LMC neuron identity specification on cell body migration. Medial LMC motor neurons are born first, and express the LIM homeodomain transcription factor Isl1. Later-born lateral LMC neurons also initially express Isl1 yet they turn it off relatively quickly, and turn on the expression of Lim1 (Tsuchida et al. 1994). At the cellular level this coincides with the migration of lateral LMC neurons, near or through the cluster of medial LMC neurons, which at this time begin to express the retinoic acid synthesis enzyme retinaldehyde dehydrogenase 2 or RALDH2. Functional experiments argue that this conjunction of lateral and medial LMC neurons is required to turn off Isl1 expression and turn on Lim1 expression, providing an example migratory behaviour in spinal motor neurons that appears to be important for the induction of a particular neuronal identity (Sockanathan and Jessell 1998).
5 Effectors of Spinal Motor Neuron Migration
5.1 Reelin Signaling
The analysis of the role of transcription factors in the specification of spinal motor neurons position certainly yielded some important molecular handles on the problem of motor neuron soma migration. These are now being linked to specific effectors of neuronal migration that mediate signals from the outside environment to the neuronal cytoskeleton. Perhaps the best-known pathway that transduces such a signal is the one responding to the extracellular matrix protein Reelin (see also Chap. 1). It is a large, secreted protein, originally identified through a mutation in its gene, which leads to a locomotor behavioural phenotype due the aberrant localisation of cerebellar neurons (D’Arcangelo et al. 1995; Hamburgh 1963b). In the cortex, Reelin is deposited in the superficial layer by Cajal-Retzius neurons (Hirotsune et al. 1995; D’Arcangelo et al. 1995) while ApoER2 and Very Low Density Lipoprotein Receptor (VLDLR) are two principal Reelin receptors that transduce the signal to the cytoskeleton through the Dab1 adaptor phosphoprotein (Hiesberger et al. 1999; Trommsdorff et al. 1999). Mice, mutant for genes encoding these components, display severe disruptions in cortical layering, and on the basis of these phenotypes, Reelin has been postulated to act as a neuronal migration stop signal (Falconer 1951; Hamburgh 1963a; Hiesberger et al. 1999; Howell et al. 1997; Sheldon et al. 1997; Tissir and Goffinet 2003; Trommsdorff et al. 1999; Jossin and Cooper 2011). The downstream targets of this pathway include the cytoskeleton-associated proteins Lissencephaly1 (LIS1) and Doublecortin (DCX) (Tissir and Goffinet 2003). Reelin signalling, and its role in cortical projection neuron migration, is described in greater detail in Chap. 1.
Reelin is also expressed in the spinal cord and is required for the normal migration of PG neurons as well as the layering of the dorsal horn laminae (Yip et al. 2000, 2003a, 2004, 2009; Villeda et al. 2006; Phelps et al. 2002). Interestingly, the altered position of PG motor neurons in Reeler mutants appears to have no bearing on their connectivity to their ganglionic targets (Yip et al. 2003b) arguing that cell body position and axon projections of motor neurons can be uncoupled. This is also paralleled in more rostral regions of the nervous system where despite being inappropriately positioned, many neurons in the Reeler mouse project their axons to appropriate targets (Caviness 1976; Caviness and Frost 1980, 1983). Interestingly, the change in spinal dorsal horn layering organisation seen in Reeler mutants does have another important functional consequence: Reeler mutants have abnormal nociceptive responses (Wang et al. 2012), although in this case it is somewhat difficult to unequivocally exclude the contribution to this effect of inappropriately-developed rostral regions of the nervous system.
The specification of LMC soma position by transcription factors expressed in specific subpopulations of spinal motor neurons raised the question of whether these effects might be mediated by the Reelin pathway. Indeed, Reelin is expressed in the ventral spinal cord, and its receptors as well as the signalling intermediate Dab1 are expressed by LMC neurons (Palmesino et al. 2010). Moreover, Reelin and Dab1 mutants display LMC soma position defects: while LMC cell bodies are found in their appropriate general ventrolateral location in the spinal cord, the lateral LMC and medial LMC neuron positions are inverted along the mediolateral axis. One intriguing explanation for this phenotype is that Dab1 is expressed differentially in these neurons, with lateral LMC neurons having higher levels of the protein, compared to medial LMC. The model that emerged from additional gain and loss of function experiments is that Dab1 expression gates the LMC neurons’ sensitivity to Reelin, such that lateral LMC neurons display high Dab1 levels and thus are sensitive to Reelin located close to the ventricular zone. In contrast, medial LMC neurons display lower levels of Dab1, and thus are relatively insensitive to Reelin. This model, which is further supported by Reelin overexpression experiments (E. Palmesino and A. Kania, unpublished), posits that Reelin is not an absolute migration stop signal (D’Arcangelo et al. 1995; Ogawa et al. 1995), but rather a repulsive or migration promoting cue for LMC motor neurons, an idea in line with previous experiments in which ectopic overexpression of Reelin rescues cortical migration defects (Magdaleno et al. 2002).
The differential Dab1 expression in medial and lateral LMC neurons suggested an intriguing possibility that these levels are controlled by the same transcription factors that control LMC soma position. Indeed, Foxp1 and Lim1 gain or loss of function result in, respectively, increase or decrease of Dab1 expression in LMC neurons, and are the first example of the control of Reelin pathway protein expression by specific transcription factors (Palmesino et al. 2010). Moreover, since the same transcription factors also control LMC axon trajectory in the limb, these observations suggest a molecular hierarchy where specific transcription factors co-ordinately control both cell body position and axon trajectory, and thus establish myotopic organisation. At this point, it is unclear whether this idea can be extended to cortical or cerebellar neurons, two populations in which Reelin signaling and migration as well as molecular differentiation have been studied.
5.2 Cadherin Signaling
This large family of proteins has been associated with many types of adhesive cell-cell interactions (Nollet et al. 2000). Within the nervous system, cadherins are selectively expressed in a combinatorial manner and thus, can be used to subdivide many neuronal populations (Suzuki et al. 1997). This property, combined with their adhesive function, endows them with the ability to control neuronal soma position, as in the case of hindbrain motor neurons (Garel et al. 2000). In the context of spinal motor neurons, type II cadherin expression is used to delineate specific motor pools, suggesting a possible role in motor pool sorting. Indeed, gain and loss of function experiments in this system, where the expression of a single cadherin protein is altered, result in inappropriate clustering of motor neurons such that motor neurons from one transcription factor-defined motor pool are mixed with those from another pool (Price et al. 2002). Additional experiments argue that cadherin-mediated control of LMC neuron migration occurs through catenins, cytoplasmic effectors that link the membrane-bound cadherins to the underlying cytoskeleton (Bello et al. 2012; Price 2012). Extending this idea further, Jessell and his colleagues used mouse genetics to block catenin function and cadherin signalling in spinal motor neurons (Demireva et al. 2011). In such mice, the LMC neuron pool and division positions, as well as those of PG neurons are scrambled, while molecular identity is maintained. Moreover, the limb axonal trajectory of such motor neurons appears normal, providing further evidence that motor neuron soma position and axon trajectory can be uncoupled through the inactivation of specific molecular effectors. Conversely, in such experiments, the link between transcriptional identity and axon trajectory is maintained.
One outstanding question raised by these experiments is how cadherins are acting to selectively position spinal motor neuron cell bodies. In order for the selective adhesion mediated by combinations of cadherins to exert its effect on soma position and thus to allow for the different combinations of cadherins to interact with each other, motor neuron cell bodies need to have an opportunity to interact with one another. For this to happen, cell bodies of motor neurons could be mobilised randomly, like particles exhibiting Brownian motion, or in a more directed fashion, perhaps away from a source of a repellent, such as Reelin. Thus the relative contribution of Reelin signaling and cadherin-mediated adhesion in the specification of motor neuron position becomes an important question. The LMC soma positioning defects in catenin inactivation mutants are apparently more severe than those found in Reelin and Dab1 mutants. It might be that in order for Reelin signaling to occur, close cell-cell contacts need to form, something that could be precluded by inhibition of cadherin signalling. Thus, Reelin signalling in spinal motor neurons, might be dependent on cadherin function, and it will be interesting to evaluate in the future if in motor neurons the intersection of Reelin and cadherin signaling occurs in a mechanistically similar manner like in cortical projection neurons (see also Chap. 1).
5.3 Eph Signaling
The Eph receptor tyrosine kinases and their ephrin ligands are also important protein families that are expressed in many developing tissues, including the nervous system. They have been implicated in cell sorting, where cells that are initially intermingled become sorted into two separate compartments and thus form a boundary (Klein 2012). One seminal observation has been that of the function of Eph receptors in the compartmentalisation of hindbrain, where molecularly distinct neurons in adjacent rhombomeres form a sharp boundary (Xu et al. 1999). Unlike cadherins, there is ample evidence of the involvement of Eph receptors and their ligands in motor axon trajectory selection (Kao et al. 2012). Thus, ephrin-Eph signaling is well positioned to influence both motor neuron soma position and axon trajectory, coordinating their myotopic organisation and raising the question whether Eph signalling is involved in spinal motor neuron migration. In mice lacking EphA, a receptor important for the correct targeting of lateral LMC axons to dorsal limb muscles, the tibialis motor neuron pool is displaced within the lumbar spinal cord, arguing that this receptor is important for the normal positioning of spinal motor neuron cell bodies (Coonan et al. 2003). Interestingly, in these animals, the tibialis motor neurons still innervate their correct muscle targets, suggesting that Eph receptors can specify LMC soma position independently of axonal trajectory. This is particularly interesting given the expression patterns of ephrins and Eph receptor proteins at the level of spinal motor neuron cell bodies, and is very suggestive of their restriction to specific motor pools (Iwamasa et al. 1999).
5.4 Other Cell Surface Effectors
Although at this point there is no evidence of their function in neuron soma migration, semaphorins and their transmembrane receptors neuropilins have elaborate expression patterns at the level of hindbrain and spinal motor neuron somata (Cohen et al. 2005; Huber et al. 2005). There is accumulating evidence that these families of proteins function in the specification of motor axon trajectory in the limb (Huber et al. 2005; Moret et al. 2007; Huettl et al. 2011; Sanyas et al. 2012). Interestingly, since semaphoring-neuropilin signaling has been previously implicated in cortical neuron migration (Chen et al. 2008), it would not be at all surprising if such signaling also impacts spinal motor neuron soma localisation, although so far no evidence of this function has been reported.
6 Organisation of Spinal Motor Neurons
The above experiments point to a clear conclusion that started to emerge following cellular level studies: spinal motor neuron somata are positioned in a stereotyped manner that is highly correlated with their axonal trajectory, and thus muscle target, as well as their dendritic arborisation. The stereotypy implied that specific molecular pathways position these neurons, as outlined in the preceding section. Moreover, cellular anatomical and structural features that are linked to soma position have a substantial impact on the function of a motor neuron, highlighting the critical nature of soma position specification. The impact of axon trajectory on function is evident: the specificity of synaptic connections is a hallmark of essentially any nervous system, thus, in order to achieve efficient locomotor behavior, motor neurons must be connected to their appropriate muscle targets (Fig. 8.2).
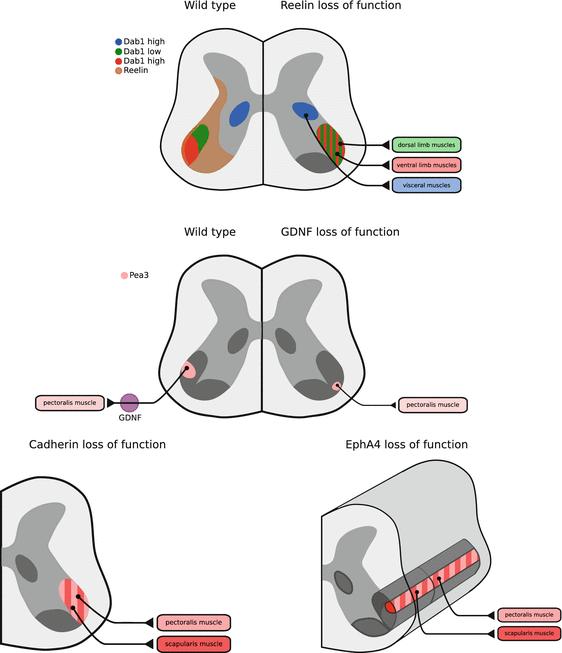

Fig. 8.2
Cell surface effectors of spinal motor neuron migration. Summary of expression of Reelin signaling pathway components, and consequence of Reelin, GDNF, Cadherin and EphA4 loss of function on spinal motor neuron soma localisation
The significance of the location of the cell body of a motor neuron has only recently been addressed in relation to its function. One obvious impact this might have is at the level of sensory-motor connectivity: sensory neurons residing in the dorsal root ganglion innervate specific muscles in the periphery, and transmit information about their proprioceptive properties directly to motor neurons innervating that particular muscle (Eccles et al. 1957). Thus, if motor neuron cell bodies are found in variable positions within the ventral horn of the spinal cord, the specificity of sensory-motor connectivity might be compromised. Indeed, emerging evidence supports this idea: in mice whose spinal motor neuron position is scrambled due to Foxp1 mutation, proprioceptive sensory axons terminate in stereotyped ventral horn positions, even if their normal motor neuron targets are not found there (Surmeli et al. 2011). This should presumably have a devastating consequence on locomotion, and reveals the existence of a sensory axon targeting mechanism that operates independently of soma position. These experiments raise a question of the consequence of molecular pathway manipulations, particularly cadherin or Reelin blockade, on sensory-motor connectivity and locomotor function.
Moreover, what about the relationship of dendritic organisation of motor neurons and their spinal cord position? The dendritic arbors of spinal motor neurons are quite complex, enabling them to integrate information arriving not only from direct proprioceptive sensory inputs, but also from spinal interneurons modulating locomotor behaviours or other sensory inputs such as nociception (Brown 1981; Rall et al. 1967). Analyses of mice mutant for the ETS transcription factor Pea3, expressed in selected spinal neuron motor pools (Lin et al. 1998), reveal that in addition to defects in pool position, the dendritic arbor of these motor neurons is also affected (Vrieseling and Arber 2006). The consequence of these changes on locomotor behaviour are unclear, however, the sensory-motor connectivity in these mutants is compromised. Given that spinal motor neurons whose cell bodies are misplaced due to cadherin signalling defects have aberrant dendritic arbors (Demireva et al. 2011), and that blocking the Reelin signal results in abnormal dendritic morphologies (Senturk et al. 2011; Rice et al. 2001), it is plausible that cell body position could potentially impact how spinal motor neurons receive and integrate the signals normally passing through axon-dendritic synapses. Another recent finding that potentially impacts how we view motor neuron soma position, functional connectivity and dendritic arborisation is the observation that pre-motor interneurons that directly synapse onto spinal motor neurons are organised in a spatially segregated manner that mirrors that of motor neuron divisions (Tripodi et al. 2011).
Another important consequence of soma position on motor neuron development and function could be at the level of their electrical coupling. There is ample evidence that in developing spinal motor neurons, connexin proteins function to maintain electrical synapses that are important for the synchronisation of electrical activity within motor pools (Fulton et al. 1980; Chang et al. 1999). Disruption of gap junctions results in decreased synchronous neuronal activity and precocious neuromuscular synapse elimination, highlighting the importance of such coupling (Personius et al. 2007). Thus, one of the consequences of inappropriate positioning of motor neuron cell bodies would be the disruption of the gap junctions that couple all motor neurons innervating a specific muscle, leading to the aberrant innervation of target muscles. Recording of neuronal activity patterns in spinal cords with disrupted myotopic organisation should reveal whether this prediction holds true.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree








