Fig. 7.1
Germinal zone migration in the developing cerebellum. (a) Cerebellar granule neuron precursors (cGNPs) migrate tangentially (horizontal arrows) within the External Granule Layer (EGL). They then transition to a radial migration mode (vertical arrows) and migrate along glial fibers through the Molecular Layer (ML) and into the Internal Granule Layer (IGL). (b) Cereballarslice cultures electroporated with CGN-specific H2B-mCherry nuclei to track neuronal migration. At postnatal day 8 (P8, 24 h post electroporation), H2B-mCherry labeled CGNs migrate tangentially through the EGL. By P10 (72 h post electroporation) most CGNs have evacuated the EGL and migrated radially into the ML and IGL
Advances in microscopy have allowed ever more detailed views of the morphology of migrating neurons in both dissociated culture and slice imaging systems. CGNs have provided a prototypic model for examination of neuronal migration, progressing from studies of fixed cells to high temporal–resolution live imaging assays of migrating cells. First, electron microscopy of the developing cerebellum in Rhesus macaques showed the migration of individual CGNs perpendicular to the surface of the brain along radial fibers later identified as Bergmann glia, with leading and trailing processes extending from their elongated soma (Rakic 1971, 1972). At the junctions of migrating CGNs, electron microscopy identified interstitial densities, or regions of the cell in which submembranous cytoskeletal elements attach to microtubules, thereby anchoring the cytoskeleton to a point at which forward force can be generated from cell-cell contacts (Gregory et al. 1988). Subsequent time-lapse imaging revealed that CGNs are highly polarized, having dynamic leading and trailing processes, while the nucleus occupies most of the somal volume. This polarity facilitates nuclear movement as a crucial aspect of saltatory CGN migration (Edmondson and Hatten 1987; Rivas and Hatten 1995; Solecki et al. 2004).
As with all neurons, the dynamic leading processes of CGNs are guided by extracellular cues but their movements are not synchronized with those of the neuronal soma (Edmondson and Hatten 1987). Polarized somal and organelle movement during CGN migration provided a foundation for understanding the basis of the saltatory movement cycle, in which the soma moves at an average rate of 33 ± 20 μm/h (Edmondson and Hatten 1987). Interestingly, forward movement of vesicles precedes somal movement, implying that specializations in cellular structures occur prior to somal movement. This concept was expanded with the observation that the centrosome enters the leading process prior to somal translocation, in what is termed the two-stroke motility cycle (Fig. 7.2) (Solecki et al. 2009). Strikingly, the saltatory timing first observed in early differential interference contrast (DIC) microscopy studies matches the two-stroke motility cycle of centrosome and soma (Solecki et al. 2009). This original observation of the mechanisms of CGN migration was expanded to apply to several other neuronal subtypes (Bellion et al. 2005; Schaar and McConnell 2005; Tsai et al. 2007; Sakakibara et al. 2013; Yanagida et al. 2012; Yang et al. 2012; Shinohara et al. 2012). Recently it has been shown a cytoplasmic dilation develops within the leading processes of subventricular-zone neurons before nuclear translocation, similar to the morphologic change seen during actin and microtubule enrichment of the leading processes of CGNs (Schaar and McConnell 2005; Rivas and Hatten 1995). Thus in vitro studies of CGN morphology provide a cellular context for understanding the large-scale migration patterns within the developing brain.
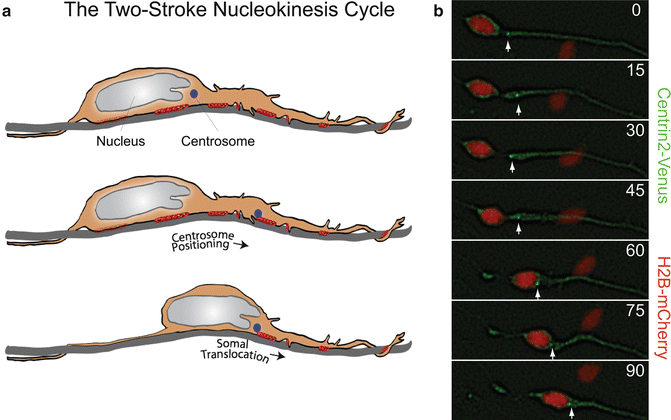

Fig. 7.2
The two-stroke nucleokinesis cycle of migrating neurons. (a) In the two stroke nucleokinesis cycle, the centrosome is positioned into the neuronal leading process before somal translocation. (b) Time-lapse imaging of a migrating CGN whose centrosome is labeled with Centrin2-Venus (green) (white arrow) and whose nucleus is labeled with H2B-mCherry (red). Centrosome positioning occurs 0 and 30 min, and somal translocation occurs between 45 and 90 min
Advances in ex vivo imaging have shown complex alterations in the morphology of migrating CGNs as they transit different environments and interact with different cell types. The shapes of radially migrating CGNs change as they pass through different layers of the developing cerebellum. The growth cone of the leading tip of migrating neurons has dynamic filopodia and lamellipodia, which are dynamic extensions that form and retract as the neuron samples its environment and moves forward (Gregory et al. 1988). In the molecular layer of the cerebellum, CGNs assume an extended shape as they move rapidly along Bergmann glia, while they assume a more rounded shape as they transiently and slowly migrate through the Purkinje cell layer. As CGNs enter the IGL, the cell body again assumes an extended shape for rapid movement independent of Bergmann glia (Komuro and Rakic 1998). Observation of tangentially migrating CGNs shows that in the EGL their velocity is dependent on their position. Their most rapid rate of tangential migration occurs in the center of the EGL, where they maintain short leading and trailing processes. As they move to the bottom of the EGL, their tangential migration velocity slows and they extend longer leading and trailing processes. CGNs slowly migrate out of the EGL upon reaching its interface with the molecular layer and begin radial migration into the molecular layer (Komuro et al. 2001). Because the multiple modes of CGN migration involve region-specific rates and morphologies, the motor systems and cytoskeletal regulation mechanisms that regulate these different types of migration are of great interest.
The cytoskeletons of migrating neurons are dynamic, changing within the different migration environments. The leading processes of migrating neurons are enriched in microtubules and actin, which extend toward a tubulin cage surrounding the nucleus (Rivas and Hatten 1995). Regulation of the microtubule cytoskeleton is a driving factor in neuronal migration. The microtubule array of migrating neurons is highly polarized, as growing microtubule “plus” tips extend into the leading process and depolymerizing “minus” ends are oriented toward the nucleus (Rakic et al. 1996). The genetics of human neuronal migration disorders further highlight that microtubule cytoskeleton and its associated motor protein dynein are regulators of neuronal migration. The cytoplasmic dynein motor protein is a polypeptide of 12 subunits, comprising two identical heavy chains that contain the AAA ATPase domains required for activity, two intermediate chains involved in cargo anchoring, and additional intermediate and light chains whose functions remain unclear (Cho and Vale 2012; Dujardin and Vallee 2002; Feng et al. 2000). Genetic analysis of lissencephaly identified mutations in the dynein adaptor protein Lissencephaly 1 (LIS1) (Reiner et al. 1993; Dujardin et al. 2003; Faulkner et al. 2000; Hirotsune et al. 1998; Smith et al. 2000). Lissencephaly also results from mutations in the Doublecortin (DCX) gene, which encodes a microtubule bundling protein and is expressed in migrating neurons (Kato and Dobyns 2003; Francis et al. 1999; Gleeson et al. 1999; Allen et al. 1998). Both LIS1 and dynein play roles in radial neuronal migration (Tsai et al. 2007; Tanaka et al. 2004; Smith et al. 2000; Shu et al. 2004) (see also Chap. 1). As more genes that participate in the regulation of neuronal migration and brain development are identified, additional genetic causes of cognitive and developmental brain disorders will be recognized.
Active migration of CGNs requires coordination between the microtubule and actin cytoskeletons and their associated motor proteins (Ridley et al. 2003). The leading processes of migrating neurons are enriched in actin, and disruption of the actin cytoskeleton with cytochalasin B is sufficient to inhibit migration, implicating actin subunit assembly in migration (Rivas and Hatten 1995; Le Clainche and Carlier 2008). Actin-based motility is dependent on the myosin family of motor proteins. Myosin II contains two heavy chains that constitute the head and tail domains of the protein and four light chains that bind to the heavy chains (Vallee et al. 2009). Phosphorylation of myosin II by myosin light chain kinase or myosin heavy chain kinase is required for ATP hydrolysis, which drives motor function (Kamm and Stull 2001; Moussavi et al. 1993). Nonmuscle myosin IIb, the main myosin expressed in the developing brain, was identified as important to neuronal migration when mutation in the motor domain of nonmuscle myosin heavy chain IIb was observed to disrupt CGN migration and cerebellar foliation (Ma et al. 2004; Vicente-Manzanares et al. 2009; Rochlin et al. 1995). Actomyosin enrichment of the leading process suggests this compartment may be the main site for actin cytoskeletal dynamics in migrating neurons (Rivas and Hatten 1995; Le Clainche and Carlier 2008).
Migrating CGNs encounter multiple microenvironments and make several types of cell-cell contact as they migrate from the EGL to the IGL (Komuro and Rakic 1998). In CGNs migrating along glial fibers, the dynamic leading process is observed to wrap around Bergmann glia, and junctional adhesion molecule (JAM)-mediated adhesions are shown to form at cell-cell contacts (Famulski et al. 2010). As the neurons encounter different cell types, their modes of migration and their adhesions change accordingly (Hatten 1990; Fishman and Hatten 1993). Astrotactin provides a receptor system for CGN migration along astroglia, and the integrin β1 receptor promotes migration along laminin fibers (Edmondson et al. 1988; Fishell and Hatten 1991; Fishman and Hatten 1993). Increased astrotactin expression, identified as a general feature of migratory cells, is noted in migratory CGNs in the EGL of the cerebellum (Zheng et al. 1996). In vitro assays of CGN migration along glial membrane- and laminin-coated fibers mirrored the saltatory nucleokinesis cycle observed in slice migration assays; however, brief, limited migration was observed on collagen and fibronectin fibers (Fishman and Hatten 1993). Individual cell surface receptors have been identified by in vitro migration assays as a requirement for neuronal migration, but it is unclear which combination of receptors is used and how they are anchored to the cytoskeleton in the different migration modes in the developing cerebellum.
Cell biology and genetic studies have created a basic framework to explore how neurons migrate from a GZ to their final laminar positions. However, several challenges remain: (1) Current migration models show inconsistencies, how will these differences be resolved? (2) How will the ever increasing array of cytoskeletal regulators be woven into an integrated model of neuronal migration? (3) What mechanisms control migration initiation and migration mode during GZ exit?
As all migrating cells are polarized (i.e., have spatially defined cytoskeletal organizations that are globally coordinated to execute complex motility programs), we will address these three major challenges by examining how polarity signaling globally organizes the neuronal cytoskeleton rather than by the reductionist approach of studying single cytoskeletal components in isolation. The best characterized cell polarity signaling molecules are the evolutionarily conserved partitioning defective (PAR) proteins (Kemphues et al. 1988). The PARD3 and PARD6 adaptor proteins form a complex containing atypical PKC and the CDC42 or Rac1 Rho GTPases (Joberty et al. 2000; Lin et al. 2000). This ternary complex is critical for tight junction formation, mitotic spindle orientation, cell migration and axon specification (Munro 2006; Barnes and Polleux 2009; Nance and Zallen 2011). This chapter will discuss the multiple roles of these proteins in neuronal migration through (1) organelle structure and movement, (2) coordination of cytoskeletal dynamics and associated motors, and (3) interaction with cell-cell focal adhesions. As studies of the PARD3/PARD6/aPKC complex (the PAR complex) progress, the individual roles of the PAR proteins in the centrosome, nucleus, actomyosin cytoskeleton, and focal adhesions are becoming clearer. PAR signaling has been shown to control the two-stroke nucleokinesis cycle of centrosome motion followed by somal translocation (Solecki et al. 2004). During the nucleokinesis cycle, the PAR complex has been shown to regulate myosin II activation and the actin cytoskeleton (Solecki et al. 2009). The role of PAR proteins and their regulation of focal adhesion turnover through the seven in absentia homolog (SIAH) E3 ubiquitin ligase (Famulski et al. 2010) has introduced PAR signaling as being regulated by protein degradation. The question of how PAR controls focal adhesions leads us to investigate how polarity complexes are related to the neuronal cytoskeleton and how these two dynamic structures control cell adhesion and migration.
Taken together, the available evidence indicates that the dynamic PAR complex plays key roles in nucleokinesis and adhesion control. We will now discuss in detail the role of PAR protein in each of these processes in the following sections of the chapter.
2 The PAR Polarity Complex and Microtubule Cytoskeletal Regulation
2.1 Cerebellar Granule Neurons Migrate with Coordinated Organelle Movements
Neurons migrate via a coordinated two-stroke motion of the centrosome and nucleus. Time-lapse imaging of actively migrating CGNs shows that in the majority of migrating neurons, forward movement of the centrosome is followed by somal translocation (Solecki et al. 2004). As centrosome movement precedes nuclear movement, models have been proposed in which the centrosome acts as a microtubule organizing center, projecting microtubules rearward to the perinuclear tubulin cage and “pulling” the nucleus forward through a dynein-mediated process. A competing model shows microtubules from the nuclear tubulin cage extending past the centrosome and anchoring in the membrane of the leading process (Higginbotham and Gleeson 2007; Tsai and Gleeson 2005). The relationship between centrosome positioning and nuclear translocation may differ among migration modes, as in vitro migration assays of CGNs identify a subset of neurons in which the nucleus overtakes the centrosome during active migration (Umeshima et al. 2007). Electron microscopy of CGNs has shown that microtubules extend from the nuclear cage forward to both the centrosome and the leading process membrane, although the anchor point for the microtubule cytoskeleton in the leading process remains unclear. As described in the next section, the PARD6 component of the PAR complex plays an important role in not only regulating the structure the tubulin cage but also the saltatory cadence of centrosome and somal motility.
2.2 PARD6 Signaling Controls Centrosome Positioning and Microtubule Dynamics
Using high temporal–resolution live imaging techniques, Solecki and colleagues (2004) demonstrated that the forward movements of the centrosome and the nucleus are tightly coordinated in migrating neurons. Photobleaching experiments showed the microtubule cytoskeleton to be highly dynamic. Overexpression of PARD6α in granule neurons inhibits neurite extension and disintegration of the perinuclear tubulin cage, showing that PARD6α controls the microtubule dynamics of migrating neurons (Solecki et al. 2004). Disruption of PARD6α signaling also uncoupled the movements of the centrosome and nucleus and prevented migration of granule neurons along Bergmann glial fibers. By using Venus-labeled PARD6α, Solecki et al. (2004) showed PARD6α to colocalize with γ-tubulin and therefore to be a component of the centrosome. PARD6α shows a relationship to centrosome structure, as overexpression of PARD6α reduced levels of centrosomal γ-tubulin (Solecki et al. 2004).
The mechanism by which PARD6 mechanistically controls centrosome positioning and migration has recently been clarified in non-neuronal systems such as epithelial cells. PARD6α siRNA disrupts the microtubule cytoskeleton in epithelial cells (Kodani et al. 2010). PARD6 is also a controlling element of the mitotic spindle, as RNAi of either PARD6α or PARD6γ causes multipolar spindle formation and mitotic failure in epithelial cells (Kodani et al. 2010; Dormoy et al. 2013). In epithelial cell wound healing assays, PARD6γ RNAi-depleted cells were unable to migrate (Dormoy et al. 2013). As overexpression of PARD6α uncouples centrosomal and nuclear movement and disrupts migration, it remains unclear whether the centrosome was acting as an organizer of polarity and migration or a reporter of cellular mechanisms that control migration in these studies.
The PAR polarity complex may play both structural and signaling roles at the centrosome in migrating neurons. In epithelial cells, PARD6α interacts with the centriolar components PCM-1 and dynactin subunit p150Glued, as shown through colocalization and immunoprecipitation studies (Kodani et al. 2010). The recruitment of PARD6α to the centrosome requires intact microtubules and dynein, as PARD6α was dispersed from the centrosome when microtubules were destabilized by nocodazole treatment and when dynein function was inhibited by overexpression of dynactin subunit p150Glued (Kodani et al. 2010; Young et al. 2000). Depletion of PARD6α by RNAi reduced microtubule-dependent recruitment of the centrosome proteins pericentrin, PCM-1, centrin, ninein, Cep170, and CPAP, showing that PARD6α promotes centrosome protein recruitment (Kodani et al. 2010). PARD6γ has been found to be a component of the mother centriole (Dormoy et al. 2013) and is required for recruitment of centrosome proteins such as PARD6α and p150Glued. Interestingly, recruitment of PARD6γ to the mother centriole is microtubule-independent and requires the C-terminus of the PARD6γ protein (Dormoy et al. 2013). These findings show that PARD6 plays both structural and recruitment roles in the centrosome. It is currently unclear where PARD6 lies in the hierarchy of centrosome protein assembly.
2.3 PARD3 Regulates Centrosome Protein Recruitment and Orientation
PARD3 plays roles other than those of PARD6 in regulating centrosomal dynamics. PARD3 associates with dynein, as shown by co-immunoprecipitation of PARD3 with dynein light intermediate chain 2 (Schmoranzer et al. 2009). Dynein is required for assembly of γ-tubulin on centrosomes (Young et al. 2000), supporting the role of the PAR complex in proper centrosome assembly (Fig. 7.3). Removal of PARD3 prevents correct centrosome positioning in relation to the nucleus in epithelial cells (Schmoranzer et al. 2009). Like PARD6, PARD3 plays roles in both directed migration and organelle positioning, and PARD3 RNAi depletion inhibits migration of epithelial cells in wound healing assays (Schmoranzer et al. 2009). Depletion of PARD3 results in increased microtubule dynamics at cell-cell contacts, showing that PARD3 plays a role in microtubule stability in epithelial cells (Schmoranzer et al. 2009). Additional studies have shown PARD3 to stabilize and bundle microtubules both in vitro and in hippocampal neurons (Chen et al. 2013). The role of the PAR complex at the centrosome, as observed in several migrating cell types, and its link to dynein, add to our understanding of regulation of the cytoskeleton and neuronal migration by the PAR polarity complex.
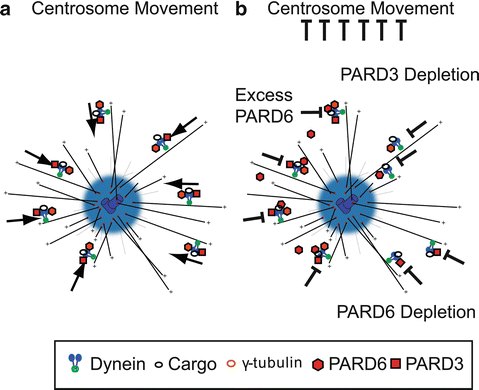

Fig. 7.3
PAR proteins and cytoplasmic Dynein directed minus-end transport. (a) Model of Dynein-directed minus-end transport of centrosome components mediated by PAR. This mechanism is responsible for proper centrosome motility. (b) Disruption of PAR protein components may result in inhibition of dynein mediated centrosome assembly and centrosome motility by PARD6 overexpression (Solecki et al. 2004), PARD6 RNAi (Kodani et al. 2010), and PARD3 depletion (Schmoranzer et al. 2009)
The PAR polarity complex and its individual components have been shown to play roles in centrosome structure, protein recruitment, and motility in addition to regulating the dynamics of the microtubule cytoskeleton. After examining the role of the PAR polarity complex in controlling migration through microtubule-based mechanisms, we will explore the role of this complex in regulating the actin cytoskeleton.
3 PAR Complex Regulation of Myosin II Motors
3.1 PARD6α Regulates Myosin II Dynamics in Migrating CGNs
Identification of PAR polarity proteins as regulators of the microtubule cytoskeleton during neuronal migration led to examination of other cytoskeletal elements in migrating neurons. While many migration studies focused on force generation by microtubule-dynein systems, Solecki and colleagues (2009) examined the role of leading-process actomyosin in migrating CGNs. Two opposing models have been proposed for the mechanism by which actomyosin contributes to force generation in neuronal migration: (1) a dynamic forward reach-and-pull model, in which leading-process actomyosin contraction pulls the neuron forward and (2) a rearward contraction model, in which actomyosin contraction at the rear of the cell pushes the migrating neuron forward (Fig. 7.4) (Trivedi and Solecki 2011; Martini and Valdeolmillos 2010; Tsai et al. 2007). Time-lapse microscopy and photobleaching/photoactivation experiments show that leading-process actin is highly dynamic in migrating neurons, and pharmacological stabilization of the actin cytoskeleton or inhibition of the myosin II motor reduces leading-process dynamics, disrupts the two-stroke nucleokinesis cycle, and halts migration of CGNs (Solecki et al. 2009). The centrosome is central to the nucleokinesis cycle, and both actin and myosin light-chain kinase were found to accumulate at the centrosome in the leading edge of migrating neurons. The importance of myosin II to neuronal migration was shown by pharmacological inhibition of the myosin II motor with blebbistatin, which halted centrosome motion and the two-stroke nucleokinesis cycle (Solecki et al. 2009). The PAR complex is a key regulator of actomyosin dynamics in the leading process. In previous studies, overexpression of PARD6α was shown to inhibit neuronal migration (Solecki et al. 2004). The same group (Solecki et al. 2009) later reported that reduced myosin II activation in cells overexpressing PARD6α was one mechanism of migration inhibition. Overexpression of PARD6α or the truncated IQ motif of PARD6α significantly reduced leading-process actin turnover in migrating CGNs (Solecki et al. 2009), showing for the first time that the PAR complex can control myosin II through direct interaction (Fig. 7.5). Co-immunoprecipitation studies revealed that full-length PARD6α binds to myosin light chain and myosin light chain kinase and that overexpression of the PARD6α IQ domain inhibits myosin light chain binding to PARD6α (Solecki et al. 2009). In other studies in C. elegans embryos, cortical flow of actin and non-muscle myosin II transported the PARD3/PARD6/aPKC complex to the anterior of the cell, maintaining polarity (Munro et al. 2004). Myosin IIb-deficient fibroblasts show polarity defects and increased levels of cytosolic PARD3 and PARD6 (Solinet et al. 2011). The mechanism of this relationship remains unclear, although it is possible that myosin II controls proper localization and stabilization of the PAR polarity complex. These results show that the PAR polarity complex regulates actomyosin contractility in the leading process of migrating neurons via PARD6α.
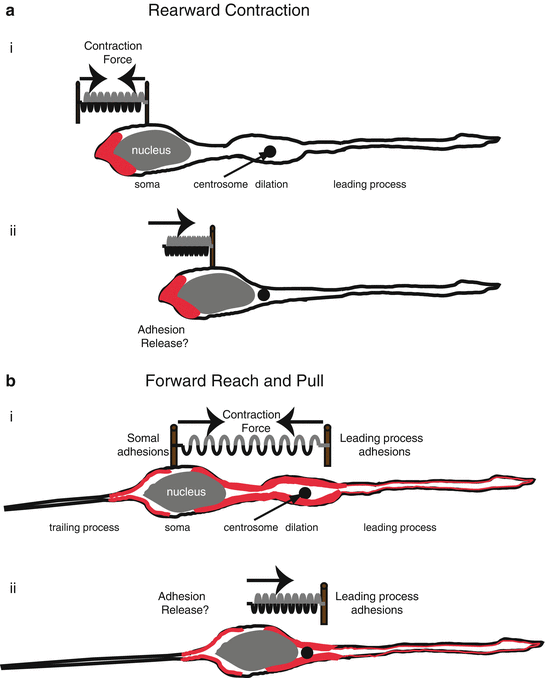
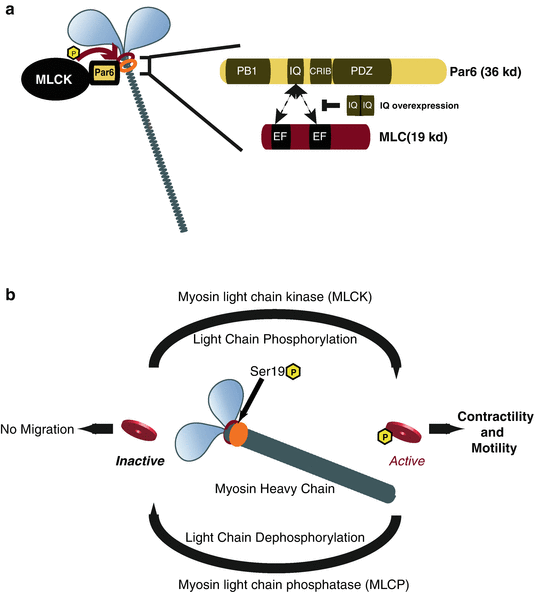

Fig. 7.4
Actomyosin pulling models for Glial-guided neuronal migration. (a) Rearward Contraction model. (i) Prior to somal movement, actomyosin (red) is heavily enriched at the cell rear. (ii) During somal movement myosin II squeezing at the rear is thought to “push” the cell body forward. (b) Reach and Pull model. (i) Prior to somal movement, actomyosin (red) is heavily enriched in the leading process from the cytoplasmic dilation to the neuronal soma. Given a muscle-like contraction of the F-actin array by myosin II, a taut spring effectively describes the forces produced when leading process and somal actomyosin anchoring (i.e., adhesions) are balanced before somal movement: one force vector points from the leading process back towards the soma whereas another force vector points from the soma towards the dilation (the future direction of somal movement). (ii) Once somal adhesions release, as described in (Gregory et al. 1988), actomyosin tension generated in the leading process primes somal movement towards the cytoplasmic dilation (Reproduced with permission of (Trivedi and Solecki 2011))

Fig. 7.5
Model of Par6α interaction with the Myosin II motor complex and the Myosin cycle. (a) Par6α binds to both MLC and MLCK, key signaling nodes regulating actomyosin contractility. Inset: The PARD6-MLC interaction may be mediated by the IQ domain of Par6α (IQ Motif (aa 104–120) = AFASNSLQRRKKGLLLRPV) and the EF hand domains of MLC. (b) Myosin contractility is dependent on Myosin Light Chain (MLC) phosphorylation by Myosin Light Chain Kinase (MLCK) at Ser19 and is required for neuronal migration. De-phosphorylation of MLC by Myosin Light Chain Phosphatase (MLCP) results in MLC inactivity and lack of myosin contractility. MCL and MLCP activity cycles Myosin contractility in migratory cells ((a) Reproduced with permission of (Solecki et al. 2009))
3.2 Actomyosin Dynamics in Migrating Neurons
Further studies examining the dynamics of actin cytoskeletal elements in CGNs buttress the importance of leading-process actin. The forward flow of actin in the leading process plays several roles important to migration. He and coworkers (2010) examined the role of cytoskeletal components and motors in vitro in distinct regions of migrating rat CGNs. In their microdissection experiments, severing the distal leading tip of migrating neurons was sufficient to inhibit somal translocation, while a dynamic leading tip contributed to somal translocation by a distance of several cell-body lengths (He et al. 2010). By micropipetting actin-destabilizing drugs (cytochalasin D, latrunculin A) or actin-stabilizing drugs (jasplakinolide) into the vicinity of the leading processes of migrating neurons, they also showed that leading-process actin dynamics are required for somal translocation. Pharmacological inhibition of leading-process actin dynamics halted somal translocation; however, when the inhibitor was concentrated in the cell body area, it did not similarly inhibit somal translocation (He et al. 2010). The same group (He et al. 2010) also compared the roles of the microtubule cytoskeleton and of actin in the leading tip and found that the microtubule-destabilizing drug nocodazole did not halt somal translocation, but rather enhanced the rate of nuclear migration. Directed inhibition of myosin II by applying blebbistatin to the leading tip of migrating CGNs halted somal translocation, while blebbistatin treatment at the rear of the cell increased the rate of nuclear migration, demonstrating the importance of the myosin II motor (He et al. 2010). These findings showed that polarized activity of myosin II plays an important role in neuronal migration.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree








